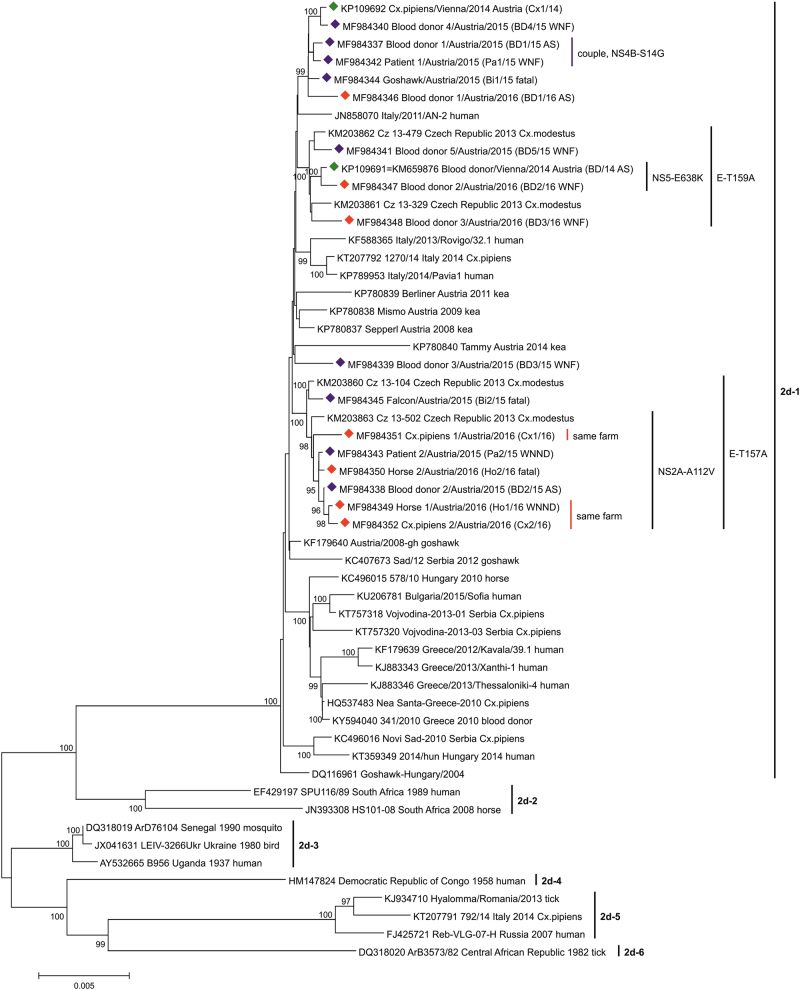
Fig. 2. Phylogenetic tree of 54 selected complete polyprotein-coding nucleotide WNV sublineage 2d sequences.
Green diamonds indicate the 2 sequences from 201418. Sixteen viruses determined in this study are marked with blue diamonds (identified during 2015) and red diamonds (2016). The six major clusters (1–6) of WNV subclade 2d are indicated by vertical bars. All Austrian viruses belong to the Central/Southern European cluster 2d-1. The GenBank accession numbers, strain names and – if not included in the strain names – geographic locations, years of identifications and host species names are indicated at the branches. For the Austrian strains, common mutations and symptoms (AS/WNF/WNND/fatal) are also depicted. Supporting bootstrap values ≥90% are displayed next to the nodes. The horizontal scale bar indicates genetic distances (here 0.5% nucleotide sequence divergence). Abbreviations used: BD, blood donor; Pa, patient; Ho, horse; Bi, bird; Cx, Culex mosquito; AS, asymptomatic; WNF, West Nile fever; WNND, West Nile neuroinvasive disease