Fig. 8.
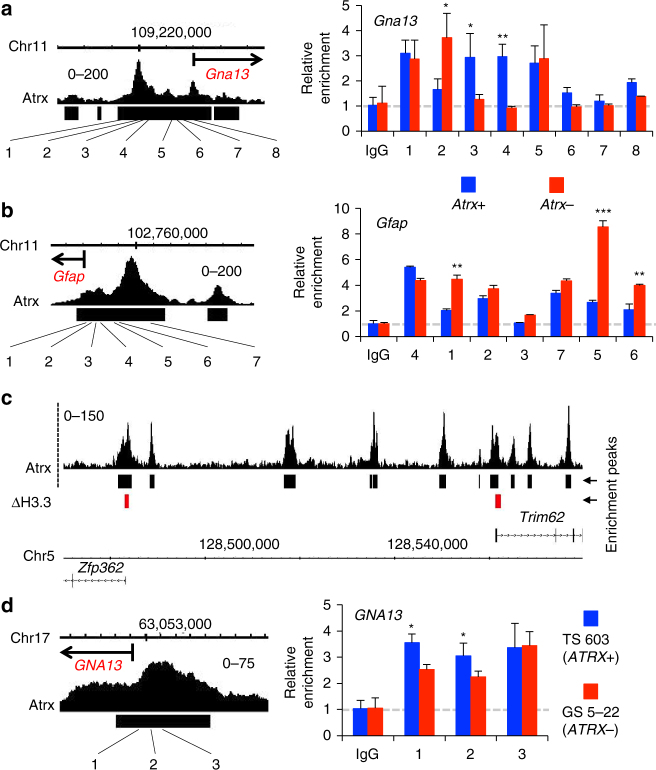
Atrx-dependent epigenetic and transcriptional alterations correlate with H3.3 composition. a, b H3.3 ChIP qPCR from Atrx-bound genomic loci approximating Gna13 and Gfap reveals differential enrichment between Atrx+ and Atrx- mNPCs at many points. Positioning of loci subjected to qPCR relative to Atrx binding peaks and open reading frames (Gna13 and Gfap) is also shown (left panels). c sample trace showing relationship of Atrx ChIP-seq peaks (black) to sites of significantly differential H3.3 composition (ΔH3.3) arising with Atrx deficiency (red). d H3.3 ChIP qPCR from Atrx-bound genomic loci approximating GNA13 reveals differential enrichment between ATRX+ (TS 603) and ATRX- (GS 5-22) human GSCs (right panels; *P < 0.05; **P < 0.001; ***P < 0.0001. In all cases, error bars reflect standard deviation and P values determined by unpaired two-tailed t-test