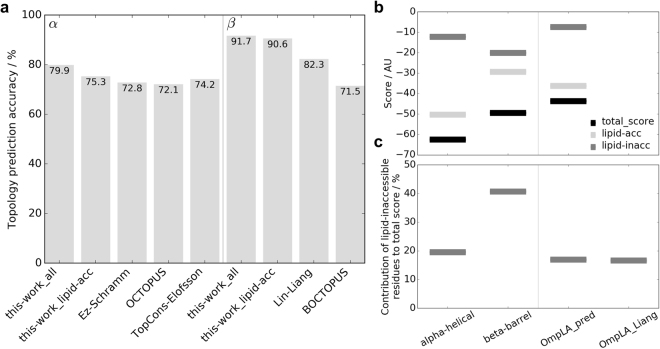
Figure 5.
(A) Prediction accuracies of different topology predictors in percent for both α-helical and β-barrel proteins. We compute prediction accuracies from residues in the membrane for both lipid-accessible and lipid-inaccessible residues (denoted ‘all’) and for lipid-accessible residues only (denoted ‘lipid-acc’) for a fair comparison to other methods. Note that OCTOPUS, TopCons and BOCTOPUS are sequence-based machine learning methods. Details about the different methods and accuracies are given in the results section. (B) Average free energy scores for the native position (position (0, 0) in Fig. 7) over all α-helical (239 proteins) and β-barrels (96 proteins) in our databases, as well as for OmpLA based on our prediction. The scores for the lipid-accessible residues (light gray) and lipid-inaccessible residues (dark gray) give rise to the total score (black). (C) Contribution of lipid-inaccessible residues to the total score. For α-helical proteins, lipid-inaccessible residues contribute on average about 19.57% to the total score – these residues are most often buried in the protein interior. For β-barrels, lipid-inaccessible residues contribute on average 40.70% to the overall score. Since lipid-inaccessible residues mostly face the aqueous pore in β-barrels, the contribution of pore-facing residues to overall insertion and stability is considerably higher than was previously suggested28. However, there is excellent agreement between the values suggested for OmpLA (right panels): Liang et al. estimated that lipid-facing residues contribute 16.67% to overall insertion, while our predicted value for OmpLA is 16.95%.