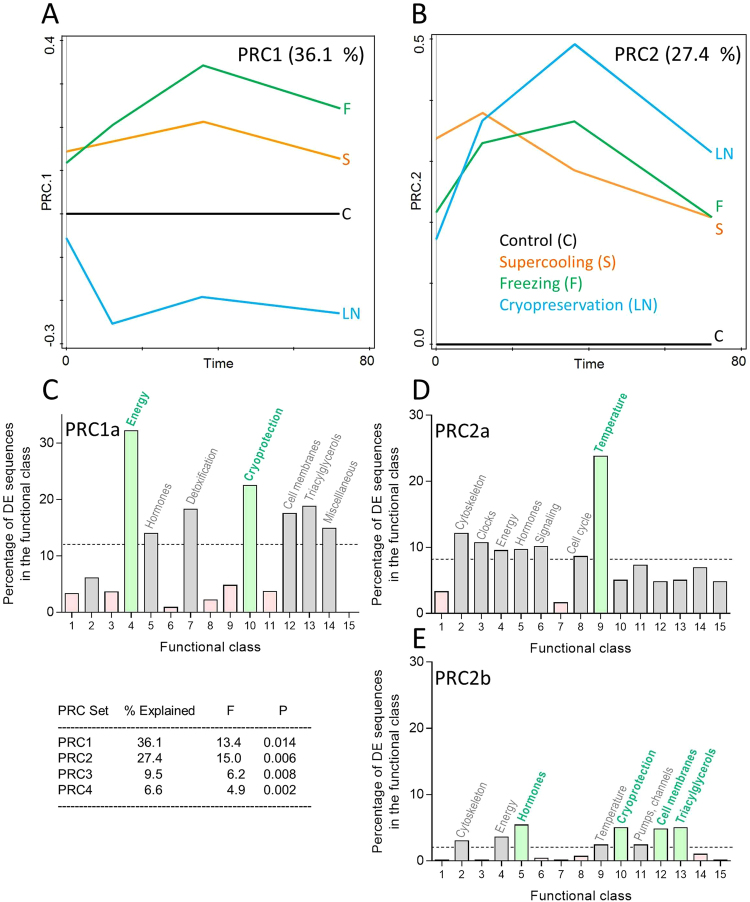
Figure 4.
Gene transcript profiles of control and cold-stressed Chymomyza costata larvae. The transcriptomics analysis was conducted using custom C. costata microarrays containing probes for 1,124 candidate gene sequences. (A,B) The principal response curve (PRC) model found two principal PRC sets showing relatively high and statistically significant proportion of variation explained by the main effect of treatment plus its interaction with time. In control larvae, C, the temporal pattern of transcriptional changes during their 3-day recovery after transfer to 18 °C is levelled to 0 and the temporal patterns of three cold-stressed treatments (S, F, LN) are normalized to the control. (C–E) The differentially expressed (DE) sequences for each PRC set (1, 2) are clusterred in functional classes and percentages of DE genes within each functional class (enrichment) is scored. See Dataset S3 for calculation of the percentages of DE sequences and for more details.