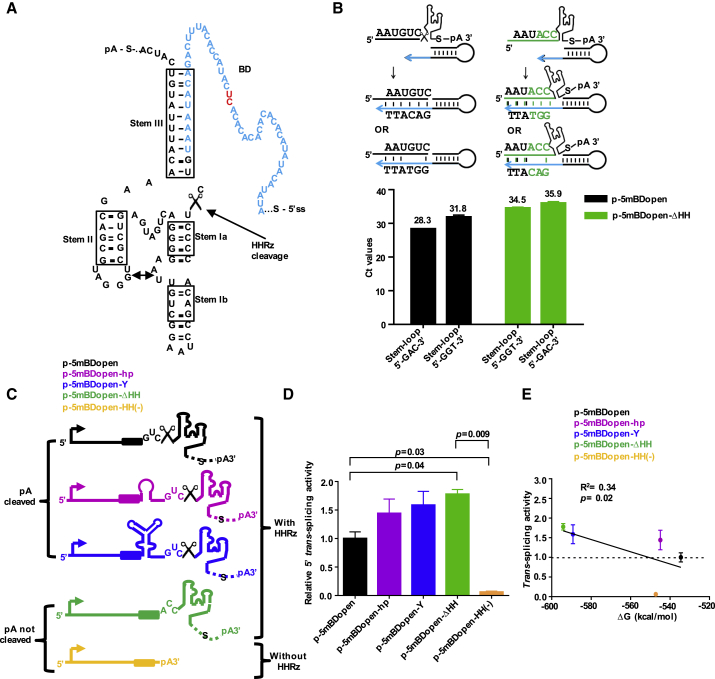
Figure 3.
Activities of 5′ ER Correlate with the Thermodynamic Stability of RNA 3′ Ends, which Can Be Processed by a HHRz
(A) RNA secondary structure as predicted by mfold of a tertiary structure-stabilized HHRz designed for cis-cleavage of tsRNA 3′ ends. The HHRz is attached downstream of the 3′ end of the target BD (blue), with indicated target mismatches (red). The cleavage site following the GUC cleavage motif is indicated by a scissor icon. Stems are highlighted in boxes; the double-headed arrow indicates tertiary structure interactions between the stem II hairpin loop and the stem I bulge. (B) Investigation of HHRz cleavage toward the active GUC or the inactive ACC cleavage motif using stem-loop primer-based real-time RT-PCR quantification of processed tsRNA 3′ ends. Blue arrow, 3′ end binding site of hairpin primer; green, mutated HHRz cleavage motif. Mean values of raw Ct values ± SEM (n = 3). (C) Schematic representation of tsRNAs for 5′ ER harboring different 3′ end stabilizing RNA secondary structures downstream of the BD and/or an active or inactive HHRz. (D) 5′ ER activities of tsRNA with different thermodynamic 3′ end stabilities relative to the parental construct p-5mBDopen harboring the active HHRz cleavage site (black). Mean ± SEM (n = 3). Significance was tested using one-way ANOVA with Tukey post hoc test. Relative RNA levels were calculated in terms of fold change (2−ΔΔCt), where ΔCt = Ct trans-spliced AFP − Ct β-Actin. (E) Correlation between relative trans-splicing activities and the Gibbs free energy (ΔG) of RNA secondary structure formation of the 5′ ER tsRNAs with different 3′ ends. Linear regression analysis was done using Prism 6 software. R2 = correlation coefficient, mean ± SD (n = 3). See also Figures S1D and S6.