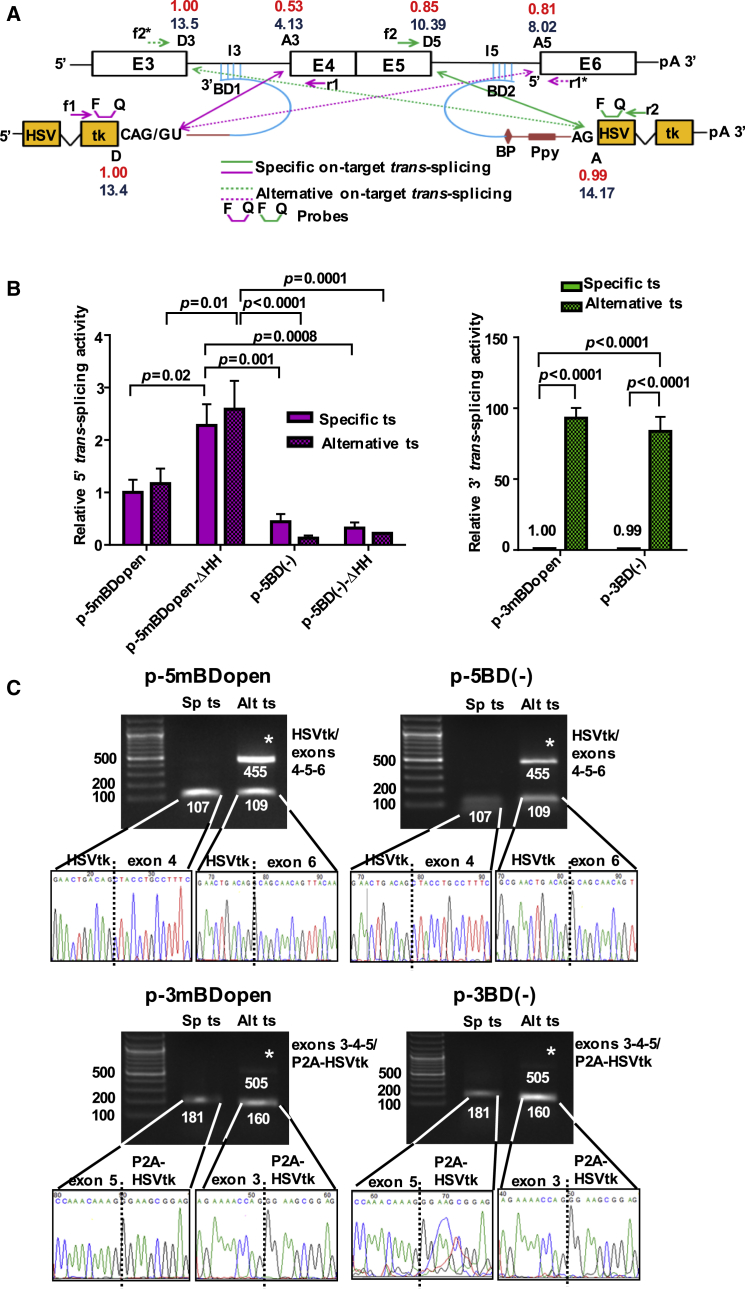
Figure 4.
Alternative On-Target trans-Splicing Is a Prevalent Event
(A) Schematic drawing depicting specific on-target trans-splicing (sp-on-ts) and alternative on-target trans-splicing (alt-on-ts) between the parental tsRNAs for 5′ and 3′ ER and the AFP mini-gene transcript. Solid arrows indicate sp-on-ts, and dashed arrows show alt-on-ts events. Numbers in red or blue denote the strengths of the donor (D) and acceptor (A) splice sites as calculated using SSP or ASSP algorithms, respectively. Positioning of primers and probes for TaqMan probe-based real-time RT-PCR detection is indicated. D and A indicate splice donor and acceptor sites of the tsRNA; D3 and D5 or A3 and A5 indicate splice D or A sites of the AFP mini-gene RNA. PCR forward primers are f1, f2*, and f2; reverse primers are r1, r1*, and r2. For further details, see the legend of Figure 1A. (B) Relative sp-on-ts and alt-on-ts activities triggered by different tsRNAs for 5′ ER (left) and 3′ ER (right) measured using real-time RT-PCR. Means ± SEM (n = 3). Significance was tested using two-way ANOVA with Bonferroni post hoc test. (C) RT-PCR (30 + 30 cycles) detection of chimeric RNA resulting from sp-on-ts and/or alt-on-ts between the overexpressed AFP mini-gene transcript and the 5′ or 3′ ER constructs via 1% agarose gel electrophoresis (upper panel) or DNA sequencing (lower panel). *Upper bands resulting from a combination of specific on-target trans-splicing and cis-splicing of the respective second intron detected with primers f1 and r1* (5′ ER) or f2* and r2 (3′ ER). See also Table S2 and Figure S14.