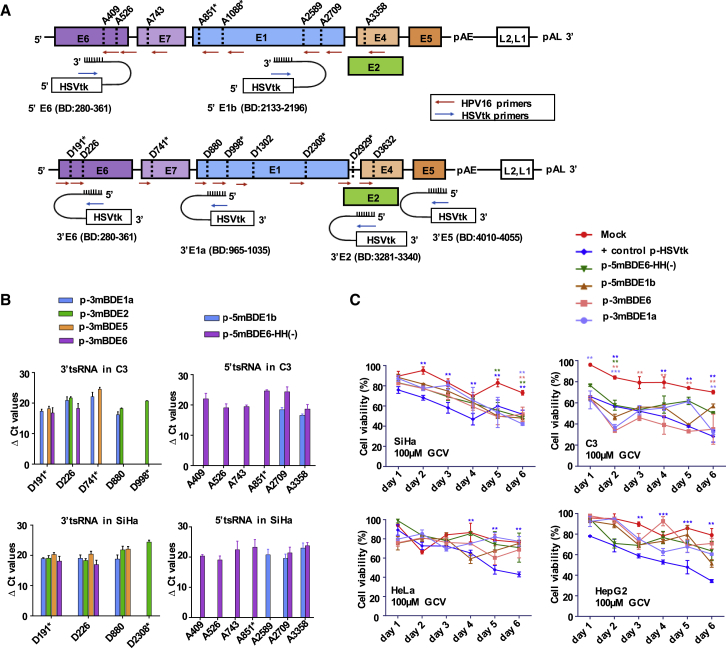
Figure 8.
Targeting of HPV-16+ Cells with trans-Splicing-Based Suicide Vectors
(A) Schematic map of the HPV-16 genome containing six early (E) and two late (L) genes with positioning of reported splice acceptor (A) or donor (D) sites, as well as splice sites predicted (*) using ASSP software. We designed two tsRNAs for 5′ ER (5mBDE6-HH(−) and 5mBDE1b) (upper panel) and four tsRNAs for 3′ ER (3mBDE6, 3mBDE1a, 3mBDE2, and 3mBDE5) (lower level). Red arrows indicate positioning of reverse (5′ ER) and forward (3′ ER) primers binding to HPV-16 transcripts; blue arrows indicate positioning of forward or reverse primers in the HSVtk region of the trans-splicing constructs. (B) Real-time RT-PCR quantification of trans-splicing between the 3′ ER (left panel) and the 5′ ER (right panel) constructs and endogenous HPV-16 transcripts. Indicated are ΔCt values normalized to β-actin levels in the HPV-16+ mouse cell line C3 (top panel) or the human cell line SiHa (bottom panel). The absence of bars for some splice donors or acceptors denotes undetermined trans-splicing. Mean ± SEM (n = 3). (C) alamarBlue cell viability assay with 3′ and 5′ ER constructs using SiHa cells and C3 cells, as well as HPV-18+ HeLa cells and HPV− HepG2 cells. Cells were treated with 100 μM GCV for 6 days. Mean ± SEM (n = 3). Significance was tested using two-way ANOVA with Bonferroni post hoc test compared to mock. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. See also Figure S13.