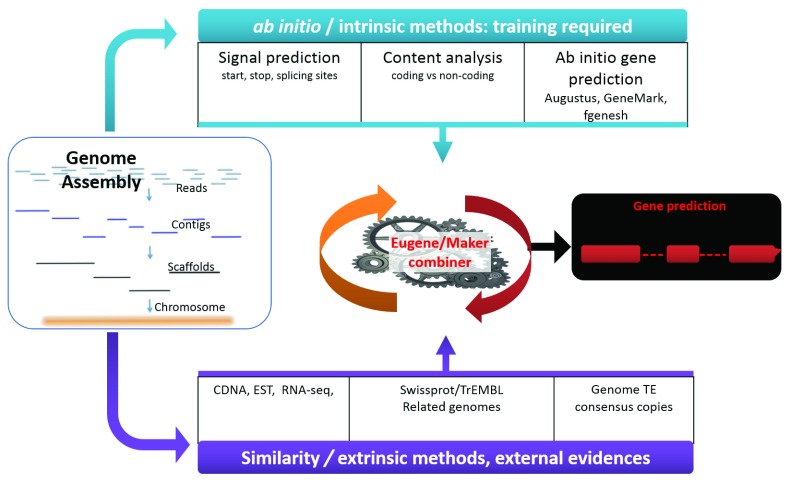
Figure 3. Simplified Illustration of a structural genome annotation using Combiners.
On the left, the diagram shows a typical assembly process. At the end of the process, scaffolds or chromosomes ready to be annotated are obtained. These scaffolds are then annotated using two different methods. The first method is called ab-initio and requires a known set of training genes. Once the ab initio tool has been trained it can be used to predict other similarly structured genes. The second similarity-based approach relies on experimental evidence such as CDSs, ESTs, or RNA-seq to build gene models. Combiners (such as Maker or Eugene) can then incorporate all of these results, eliminate incongruences, and present gene models best supported by all methods.