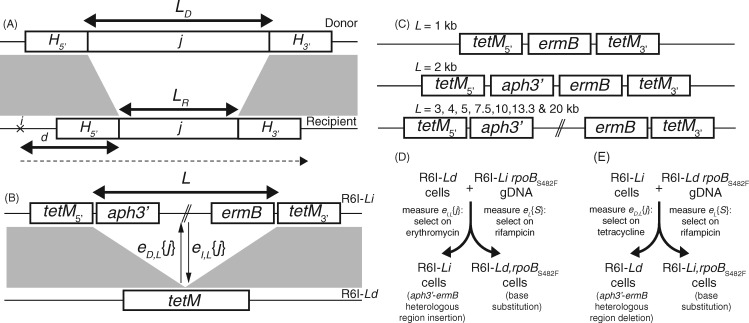
Fig. 1.
Exchange of heterologous regions through homologous recombination. (A) Description of the recombination process. Donor and recipient DNA sequences are shown with gray bands linking regions of sequence similarity, separated by a central heterologous locus j, of length LD in the donor and LR in the recipient. The minimum lengths of the flanking homologous arms necessary for exchange through homologous recombination, H5′ and H3′, are shown on either side. The dashed line indicates an homologous recombination initiating at position i, d bases upstream of j. (B) Genotypes used in the experimental system, displayed as in (A). The R6I-Li genotype had either ermB, or ermB and aph3′, inserted between two halves of the tetM gene. The R6I-Ld genotype had an intact tetM gene, with all intervening sequence in the complementary R6I-Li genotype removed. Rifampicin-resistant derivatives of both were generated through transformation with an rpoB allele encoding an S482F substitution. (C) Structure of the R6I-Li genotypes. For L = 1 kb, the insert was a single ermB gene; for L = 2 kb, both ermB and aph3′ were inserted within tetM; and for L ≥ 3 kb, these genes flanked nonessential DNA to generate constructs with the specified lengths. (D) Assaying insertion of heterology through transformation. Each R6I-Ld genotype was transformed with genomic DNA from the complementary R6I-Li rpoBS482F genotype. Insertion of heterology (eI,L{j}) was inferred from counting erythromycin-resistant colonies, and acquisition of SNPs (eL{S}) was inferred from counting rifampicin-resistant colonies. (E) Assaying deletion of heterology through transformation. Each R6I-Li genotype was transformed with genomic DNA from the complementary R6I-Ld rpoBS482F genotype. Deletion of heterology (eD,L{j}) was inferred from counting tetracycline-resistant colonies, and acquisition of SNPs (eL{S}) was inferred from counting rifampicin-resistant colonies.