Abstract
Objectives
JIA is an autoimmune disease involving disturbed T-cell homeostasis, marked by highly activated effector T cells. Autophagy, a lysosomal degradation pathway, is crucial for maintaining cellular homeostasis by regulating the survival, differentiation and function of a large variety of cells, including T cells. The aim of this study was to examine the rate of autophagy in JIA T cells and to investigate the effect of inhibition of autophagy on the inflammatory phenotype of JIA T cells.
Methods
Autophagy-related gene expression was analysed in CD4+ T cells from the SF of JIA patients and healthy controls using RNA sequencing. Autophagy was measured by flow cytometry and western blot. The effect of inhibition of autophagy, using HCQ, on the cellular activation status was analysed using flow cytometry and multiplex immunoassay.
Results
Autophagy was increased in T cells derived from the site of inflammation compared with cells from the peripheral blood of patients and healthy controls. This increase in autophagy was not induced by JIA SF, but is more likely to be the result of increased cellular activation. Inhibition of autophagy reduced proliferation, cytokine production and activation marker expression of JIA SF-derived CD4+ T cells.
Conclusion
These data indicate that autophagy is increased in JIA SF-derived T cells and that targeting autophagy could be a promising therapeutic strategy to restore the disrupted T-cell homeostasis in JIA.
Keywords: juvenile idiopathic arthritis, synovial fluid, autoimmunity, T cells, autophagy, autoimmune diseases
Rheumatology key messages
Autophagy is increased in JIA SF T cells.
Inhibition of autophagy impairs the inflammatory phenotype of JIA SF T cells.
Introduction
JIA describes a heterogeneous group of autoimmune conditions, characterized by chronic arthritis with an unknown cause and onset before the age of 16 years. Although there are several forms, two main forms of JIA are recognized based on the number of joints affected: oligo- and polyarticular JIA [1]. Inflammation in the joint SF is characterized by an increase in autoreactive, highly activated effector T cells and impaired control by Treg, but the exact disease mechanism remains unclear [2].
Autophagy is a catabolic process essential for maintaining cellular homeostasis, including T-cell homeostasis [3, 4]. Conserved in evolution, it involves the sequestration of cytoplasmic components by formation of double-membrane vesicles and subsequent delivery to the lysosome for degradation. Autophagy has been linked to T-cell activation, differentiation and survival [5–7]. There are several indications that autophagy may play a role in autoimmunity, as single nucleotide polymorphisms in autophagy-related genes are correlated with susceptibility to Crohn’s disease and SLE [8, 9]. In addition, RA serum, SLE serum and SLE purified auto-antibodies can promote autophagy in healthy control (HC) T cells [10]. Furthermore, autophagy is increased in CD4+ T cells obtained from the peripheral blood (PB) of RA patients [11].
So far, autophagy has not been studied in JIA and particularly not in primary cells derived from the site of inflammation. Given that T-cell homeostasis is disturbed in JIA and autophagy is important for T-cell homeostasis and is linked to autoimmunity, we hypothesized that autophagy is increased in JIA patient-derived T cells. Furthermore, as T-cell homeostasis is predominantly disturbed at the site of inflammation, we tested whether inflammatory mediators in SF can induce autophagy. To examine whether autophagy contributes to the inflammatory phenotype of JIA T cells, we assessed the effect of inhibition of autophagy on the activation status of JIA-derived T cells.
Methods
Sample collection and cell culture
Samples were collected, isolated and cultured as described previously [12]. Thirteen oligoarticular, eight extended oligoarticular and three polyarticular JIA patients were included in this study. All patients had active disease and underwent therapeutic joint aspiration at the time of sampling. Patients were between 5 and 18 years of age and were either untreated or treated with MTX or TNF blockers, or both at the time of inclusion. Characteristics of patient samples used in this study are outlined in supplementary Table S1, available at Rheumatology Online. Thirty-two anonymous volunteers, between 18 and 65 years old, were included as HCs. The study was approved by the Institutional Review Board of the University Medical Center Utrecht and performed according to the principles expressed in the Declaration of Helsinki. Informed consent was obtained from all patients either directly or from parents or guardians when the patients were younger than age 12 years. To obtain cell-free plasma and SF, samples were centrifuged; supernatants were collected and stored at −80 °C. Where indicated, cells were stimulated with 1 µg/ml plate-bound α-CD3 (eBioscience, San Diego, CA, USA OKT3) or cultured with HCQ (Acros Organics, Morris Plains, NJ, USA), IL-6 (BD Biosciences, San Jose, CA, USA) or TNF-α (Miltenyi, Auburn, CA, USA).
Analysis of autophagy-related genes
RNA-sequencing data from HC and JIA CD4+CD45RO+ T cells (GSE71595) were analysed for autophagy-related genes [12, 13]. Autophagy-related genes were identified via the human autophagy database (available at http://autophagy.lu/).
FACS
Autophagy was analysed using the Cyto-ID autophagy detection kit (Enzo Life Sciences, Farmingdale, NY, USA). Cells were cultured with or without HCQ, washed twice and stained with Cyto-ID (1:500) for 25 min at 37 °C. Autophagy was determined by the relative mean fluorescence intensity (MFI) Cyto-ID, that is, the difference in MFI Cyto-ID between cells treated with or without HCQ.
Apoptosis was analysed using the Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s protocol. Apoptotic cells were defined as Annexin V+.
To detect intracellular cytokine production, cultured cells were stimulated for 4 h with phorbol 12-myristate 13-acetate (20 ng/ml; Sigma, St. Louis, MO, USA) and ionomycin (1 µg/ml; Calbiochem, San Diego, CA, USA), with monensin (1:1500; BD Biosciences) for the last 3.5 h. Cells were washed twice in FACS buffer [PBS with 2% Fetal Calf Serum (Invitrogen, Waltham, MA, USA) and 0.1% sodium azide (Sigma-Aldrich, St. Louis, MO, USA)] and subsequently stained with surface antibodies. Then, cells were washed twice in FACS buffer, fixed and permeabilized (eBioscience; according to the manufacturer’s instructions) and stained with cytokine antibodies.
Western blot
Western blot was performed as described previously [11]. In short, CD4+ T cells were isolated using Biotin Human CD4+ T lymphocyte Enrichment Set-DM (BD IMag, San Jose, CA, USA) according to the manufacturer’s protocol and lysed in Laemmli buffer (0.12 M Tris–HCl, pH 6.8, 4% SDS, 20% glycerol, 0.05 µg/µl Bromophenol Blue and 35 mM β-mercaptoethanol). Samples were separated by SDS–PAGE, transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA), probed with mouse anti-LC3 (Nanotools, Teningen, Germany, 5F10) and goat anti-actin (Santa Cruz, CA, USA sc-1616), and analysed using enhanced chemiluminescence (GE Healthcare, Pittsburgh, PA, USA).
Cell proliferation
Before culture, cells were labelled with 2 µM CellTrace Violet (Invitrogen) for 7 min at 37 °C. Labelling was blocked by adding 10 volumes cold serum. The MFI of CellTrace Violet is inversely correlated with proliferation, that is, higher CellTrace Violet MFI means less proliferation.
Multiplex immunoassay
Supernatant derived from JIA SF mononuclear cells (SFMC) cultured for 4 days was collected and stored at −80 °C. Within 1 month, cytokine concentrations were measured using Luminex technology as previously described [12].
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6.0, San Diego, CA, USA. Paired samples were analysed using Student’s paired t-test, and for multiple groups statistical comparison was performed using repeated-measures analysis of variance followed by post hoc testing with Sidak’s procedure for multiple testing.
Results
Autophagy is increased in JIA SF-derived T cells compared with JIA PB-derived T cells
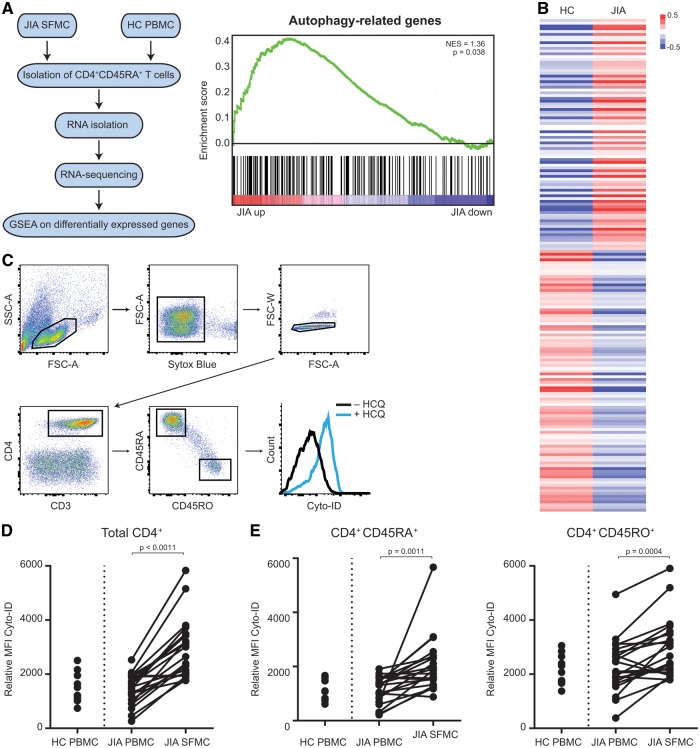
To examine whether autophagy might be affected in JIA T cells, the expression of autophagy-related genes in HC and JIA SF-derived CD4+CD45RO+ T cells was analysed using RNA-seq (Fig. 1A) [12]. Autophagy-related genes were significantly enriched within genes that are upregulated in JIA compared with HC, indicating that autophagy is disturbed in JIA T cells (Fig. 1A and B and supplementary Table S2, available at Rheumatology Online). To validate that autophagy is increased in JIA T cells, we determined autophagy in HC PB mononuclear cells (PBMC) and JIA PBMC and SFMC. To determine the rate of autophagy, that is, autophagic flux, we cultured the cells with and without HCQ. HCQ affects lysosomal acidification, hence inhibiting degradation of autophagosomes, resulting in the accumulation of autophago(lyso)somes. Cyto-ID, a cationic amphiphilic tracer dye, specifically recognizes autophago(lyso)somes and can be quantified using flow cytometry [14]. The difference in MFI Cyto-ID of cells treated with and without HCQ was used to measure the autophagic flux (Fig. 1C). This demonstrated that autophagy was significantly increased in SF-derived CD4+ T cells compared with paired PB-derived CD4+ T cells (Fig. 1D). Similar observations were made for CD8+ T cells (data not shown). The detected difference was not related to disease activity, disease subtype or treatment (supplementary Fig. S1A–C, available at Rheumatology Online). Given that autophagy was demonstrated to be increased in memory/effector T cells (CD4+CD45RO+) compared with naïve T cells (CD4+CD45RA+), and memory/effector T cells are present in large numbers in the inflamed synovium, autophagy was analysed in each subset (Fig. 1E and supplementary Fig. S1D, available at Rheumatology Online) [15]. In both subsets, a significant increase was detected in SF-derived T cells compared with PB-derived T cells, indicating that the increase in autophagy is not merely a reflection of the increased amount of memory/effector T cells in the synovium.
Fig. 1.
Autophagy is increased in JIA SF-derived CD4+ T cells compared with JIA peripheral blood-derived T cells
(A) Schematic overview of experimental set-up of RNA sequencing. (B) Gene Set Enrichment Analysis of autophagy-related genes. (C) Average log2 fold change of autophagy-related genes in HC and JIA CD4+CD45RO+ T cells. (D) Gating strategy used to analyse autophagy levels in CD4+ T cells. (E) Relative MFI Cyto-ID of CD4+ T cells in HC PBMC (n = 11), JIA PBMC and SFMC (n = 21, paired) cultured for 16 h with or without 20 µM HCQ. (F) Relative MFI Cyto-ID of CD4+CD45RA+ and CD4+CD45RO+ T cells in HC PBMC, JIA PBMC and SFMC cultured with or without 20 µM HCQ. HC: healthy controls; MFI: mean fluorescence intensity; PBMC: peripheral blood mononuclear cells; SFMC: SF mononuclear cells.
Autophagy is not induced by the SF, and inhibition of autophagy impairs the inflammatory phenotype of JIA SF-derived CD4+ T cells
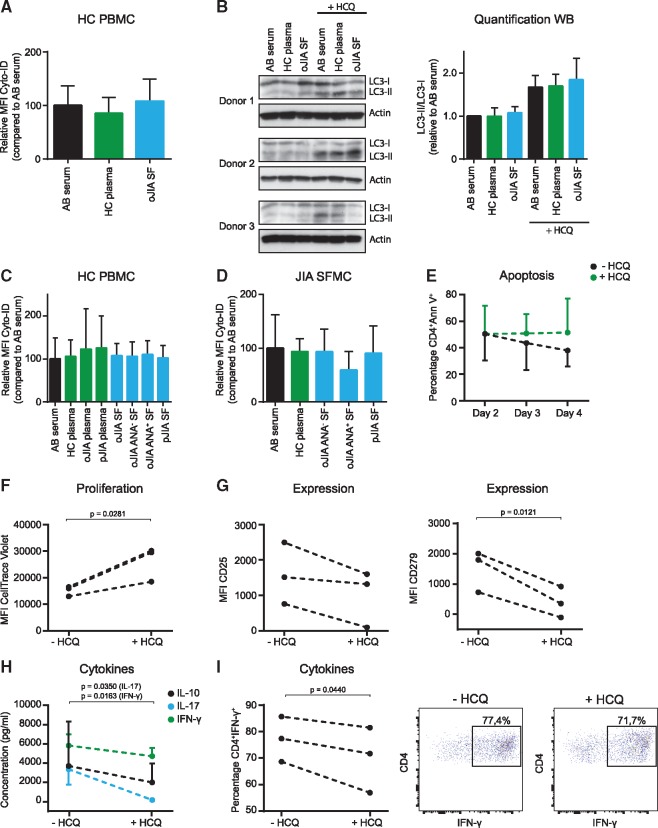
As autophagy is increased in cells derived from the synovium, we aimed to determine whether inflammatory mediators in the SF can induce autophagy. HC PBMC were cultured with either AB serum, HC plasma or pooled oligoarticular JIA SF samples, and the autophagic flux was determined (Fig. 2A). We did not detect a consistent effect of SF on autophagy in these conditions or with additional α-CD3 stimulation (data not shown) or using western blot as a technique to assess autophagy (Fig. 2B). To validate these data, plasma and SF from different JIA subtypes, discriminated according to the presence of ANA, were tested. Autophagy was not increased upon culture with these samples in either HC (Fig. 2C) or JIA SF-derived T cells (Fig. 2D). Furthermore, the effect of individual cytokines, present within the SF on autophagy, was analysed. Neither TNF-α nor IL-6 affected autophagy (supplementary Fig. S2A, available at Rheumatology Online).
Fig. 2.
Autophagy inhibition impairs the inflammatory phenotype of JIA SF-derived CD4+ T cells
(A) Relative MFI Cyto-ID of CD4+ T cells (n = 13) cultured for 16 h with or without 2.5 µM HCQ. (B) Western blot of HC CD4+ T cells (n = 3) cultured for 16 h with or without 2.5 µM HCQ, α-CD3 stimulated. (C) Relative MFI Cyto-ID of CD4+ T cells (n = 4) cultured for 16 h with or without 2.5 µM HCQ. (D) Relative Cyto-ID MFI of CD4+ T cells (n = 5) cultured for 16 h with or without 2.5 µM HCQ. (E–I) JIA SFMC (n = 3) were cultured with or without 20 µM HCQ, α-CD3 stimulated, and the percentage CD4+Annexin V+ T cells (E), CellTrace Violet MFI of CD4+ T cells (F), MFI of activation markers on CD4+ T cells (G), supernatant cytokine concentrations (H) and percentage of CD4+IFN-γ+ T cells (I) were measured. HC: healthy controls; MFI: mean fluorescence intensity; PBMC: peripheral blood mononuclear cells; SFMC: SF mononuclear cells.
Next, we wondered what the effect is of increased autophagy on the cellular activation status of JIA SF-derived T cells. Given that highly activated effector T cells are a key feature of JIA and that autophagy is involved in T cell activation, we hypothesized that autophagy contributes to the inflammatory phenotype of SF-derived CD4+ T cells [6]. To test this, we cultured activated SFMC for 4 days with HCQ, to inhibit autophagy, and determined cellular activation status by flow cytometry. HCQ treatment resulted, as expected, in increased Cyto-ID levels (supplementary Fig. S2B, available at Rheumatology Online) and did not affect apoptosis (Fig. 2E). In contrast, proliferation and expression of the activation markers CD25 and CD279 was decreased upon HCQ treatment (Fig. 2F and G). Furthermore, there was a significant decrease in cytokine production, measured both in the supernatant and intracellularly (Fig. 2H and I).
Altogether, these data demonstrate that culturing HC and JIA T cells in the presence of SF does not induce autophagy and that autophagy contributes to the inflammatory phenotype of SF-derived CD4+ T cells.
Discussion
Autophagy has recently been implicated as playing a role in numerous autoimmune diseases [4]. Here, we studied autophagy in T cells of JIA patients and observed that autophagy is increased in T cells obtained from inflamed joints.
It remains to be determined whether the increase in autophagy at the site of inflammation is specific for JIA. Given that T-cell activation induces autophagy, the increase in autophagy in JIA T cells could be a consequence of their inflammatory phenotype [5, 6, 15]. Therefore, other diseases characterized by hyperactivated T cells will probably display a similar phenotype. For various rheumatic diseases, it has been demonstrated that autophagy is affected in disease-associated (immune) cells [16]. However, the alterations at the autophagy level differ from one disease and one cell type to another, making it difficult to pinpoint the exact role of autophagy in disease pathogenesis. This might also explain why, in contrast to what has been reported for SLE and RA serum, we did not observe that JIA SF induces autophagy in HC CD4+ T cells [10]. For example, in contrast to JIA, in RA autophagy is increased in PB-derived T cells compared with HC cells, and in SLE autophagy is increased in naïve CD4+ T cells, but not in memory or CD8+ T cells [10, 11]. This suggests that autophagy may have distinct roles within different autoimmune diseases and that this might be related to differences in pathology and localization of the disease [10]. Furthermore, it is important to take into account that, apart from the inflammatory mediators present in the SF, there are many more aspects present at the site of inflammation, such as non-soluble factors and cell–cell contact (e.g. with synovial fibroblasts), that can induce immune activation and thereby affect autophagy.
Additionally, it remains unclear whether increased autophagy is a cause or a consequence of disease pathology. Our data suggest that autophagy may be necessary to increase the intracellular nutrient supply to meet the metabolic demand of activated T cells, thus pointing to a secondary role for autophagy in inflammation. However, autophagy is also described as being important for self-tolerance by regulating MHC class II antigen presentation in the thymus, suggesting that deregulated autophagy might also play a role in the initiation of autoimmunity [17].
Here, we chose to measure autophagy using western blot for LC3-I/LC3-II and flow cytometry with Cyto-ID. The advantage of flow cytometry is the relatively small amount of cells needed and the medium to high throughput of this technique. Alternative methods to measure autophagy are, for instance, SQSTM1/p62 turnover and LC3-GFP fluorescence microscopy [18]. The latter technique also allows visualization of the size of the autophagosomes, but requires cell transfection and is thus difficult to apply to primary, patient-derived cells. In the present study, we used HCQ to block autophagy. However, as HCQ affects lysosomal acidification, other lysosome-dependent processes could be affected as well [19]. To elucidate whether the effect of HCQ on the cellular activation status is primarily caused by inhibition of autophagy, genetic manipulation of autophagy-related genes could be useful.
In conclusion, we demonstrated that autophagy is increased in synovial JIA T cells. This increased autophagy is not induced by the inflammatory environment, but rather seems to be related to increased immune activation. This can be decreased by inhibiting autophagy using HCQ, a compound that is already being used for the treatment of various diseases [16]. Therefore, inhibition of autophagy might be a promising therapeutic approach to target activated, autoreactive T cells and restore the disrupted T-cell homeostasis in JIA.
Supplementary Material
Acknowledgements
We thank the Luminex core facility in the University Medical Center Utrecht, The Netherlands, for determining cytokine concentrations. Contributions: J.G.C.P., N.d.G., M.L., S.A., S.d.R. and J.v.L. designed experiments, analysed and interpreted data. J.G.C.P. and N.d.G. performed experiments. S.d.R. provided patient material. J.G.C.P., S.d.R. and J.v.L. wrote the paper. J.v.L. and S.d.R supervised the study and contributed equally to this work.
Funding: This work was supported by the Netherlands Organization for Scientific Research [grant number 022.004.018], the Dutch Arthritis Foundation [grant number 14-3-201], National Institutes of Health [grant number AG007996], National Medical Research Council (NMRC) [grant numbers NMRC/STaR/020/2013, NMRC/MOHIAFCAT2/005/2015, MOHIAFCAT2001, CIRg13nov032 and NMRC MOHIAFCAT1-6003], Duke-National University of Singapore and Biomedical Research Council [SPF2014/005].
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology Online.
References
- 1. Petty RE, Southwood TR, Manners P. et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol 2004;31:390–2. [PubMed] [Google Scholar]
- 2. Prakken B, Albani S, Martini A.. Juvenile idiopathic arthritis. Lancet 2011;377:2138–49. [DOI] [PubMed] [Google Scholar]
- 3. Klionsky DJ, Emr SD.. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levine B, Kroemer G.. Autophagy in the pathogenesis of disease. Cell 2008;132:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pua HH, He Y-W.. Autophagy and lymphocyte homeostasis. Curr Top Microbiol Immunol 2009;335:85–105. [DOI] [PubMed] [Google Scholar]
- 6. Hubbard VM, Valdor R, Patel B. et al. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol 2010;185:7349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bronietzki AW, Schuster M, Schmitz I.. Autophagy in T-cell development, activation and differentiation. Immunol Cell Biol 2015;93:25–34. [DOI] [PubMed] [Google Scholar]
- 8. Barrett JC, Hansoul S, Nicolae DL. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gateva V, Sandling JK, Hom G. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 2009;41:1228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alessandri C, Barbati C, Vacirca D. et al. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J 2012;26:4722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Loosdregt J, Rossetti M, Spreafico R. et al. Increased autophagy in CD4+ T cells of rheumatoid arthritis patients results in T cell hyperactivation and apoptosis resistance. Eur J Immunol 2016;46:2862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peeters JGC, Vervoort SJ, Tan SC. et al. Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep 2015;12:1986–96. [DOI] [PubMed] [Google Scholar]
- 13. Peeters J, Vervoort S, Mijnheer G. et al. Autoimmune disease-associated gene expression is reduced by BET-inhibition. Genom Data 2015;7:14–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oeste CL, Seco E, Patton WF, Boya P, Pérez-Sala D.. Interactions between autophagic and endo-lysosomal markers in endothelial cells. Histochem Cell Biol 2013;139:659–70. [DOI] [PubMed] [Google Scholar]
- 15. van Loosdregt J, Spreafico R, Rossetti M. et al. Hydroxychloroquine preferentially induces apoptosis of CD45RO+ effector T cells by inhibiting autophagy: a possible mechanism for therapeutic modulation of T cells. J Allergy Clin Immunol 2013;131:1443–6.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rockel JS, Kapoor M.. Autophagy: controlling cell fate in rheumatic diseases. Nat Rev Rheumatol 2016;12:517–31. [DOI] [PubMed] [Google Scholar]
- 17. Münz C. Antigen processing for MHC Class II presentation via autophagy. Front Immunol 2012;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klionsky DJ, Abdelmohsen K, Abe A. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mackenzie AH. Pharmacologic actions of 4-aminoquinoline compounds. Am J Med 1983;75(1A):5–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.