Fig. 1.
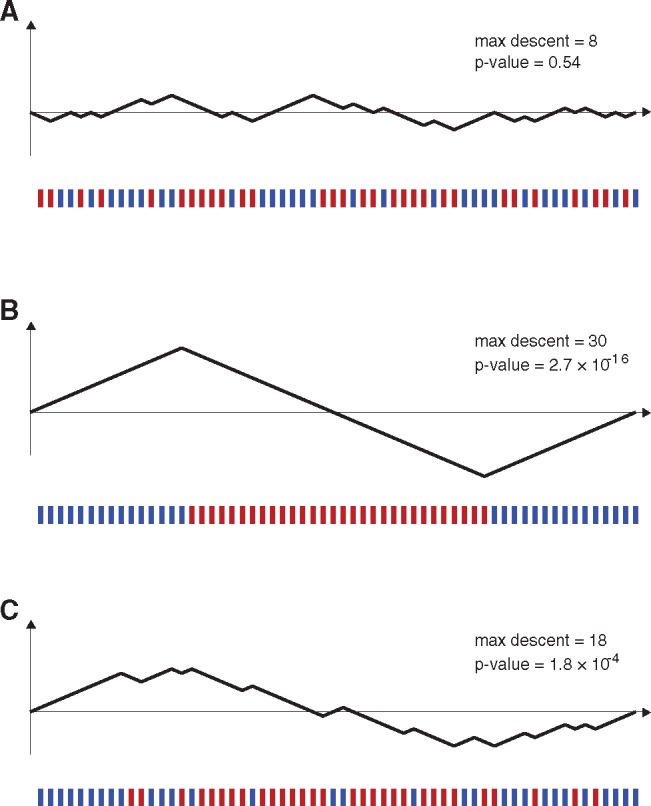
Relationship between ordering of informative sites along a genome and a hypergeometric random walk. Below each set of axes, the 30 red bars and 30 blue bars show positions on a genome (informative sites) where a putative recombinant sequence is identical to parent P but different from parent Q (blue bars), or identical to parent Q but different from parent P (red bars). Each blue site can be mapped to an up-step in a random walk and each red site can be mapped to a down-step in a random walk, and there is a one-to-one correspondence between the space of informative-site arrangements and the space of hypergeometric random walks. (A) A random arrangement of informative sites, which does not visually suggest that the sequence is a mosaic of putative parents P and Q. The arrangement of sites maps to a random walk which stays fairly close to the horizontal axis. This walk’s maximum descent is eight steps, and ∼54% of HGRWs with 30 up-steps and 30 down-steps have a maximum descent of eight steps or greater. (B) A nonrandom arrangement of informative sites that clearly suggests that the candidate sequence is a mosaic of the two parental sequences P and Q. The probability of all the red sites appearing consecutively is 31! × 30!/60! which is 2.62 × 10−16. (C) An arrangement of red sites and blue sites that suggests the red sites may be clustered in the middle. When mapping the site arrangement to a hypergeometric random walk, the random walk has a maximum descent of 18 steps. The P value for a maximum descent of 18 steps cannot be written down in closed form but can be calculated from recursion (4). The P value for this maximum descent and for this arrangement of informative sites is 1.8 × 10−4.
