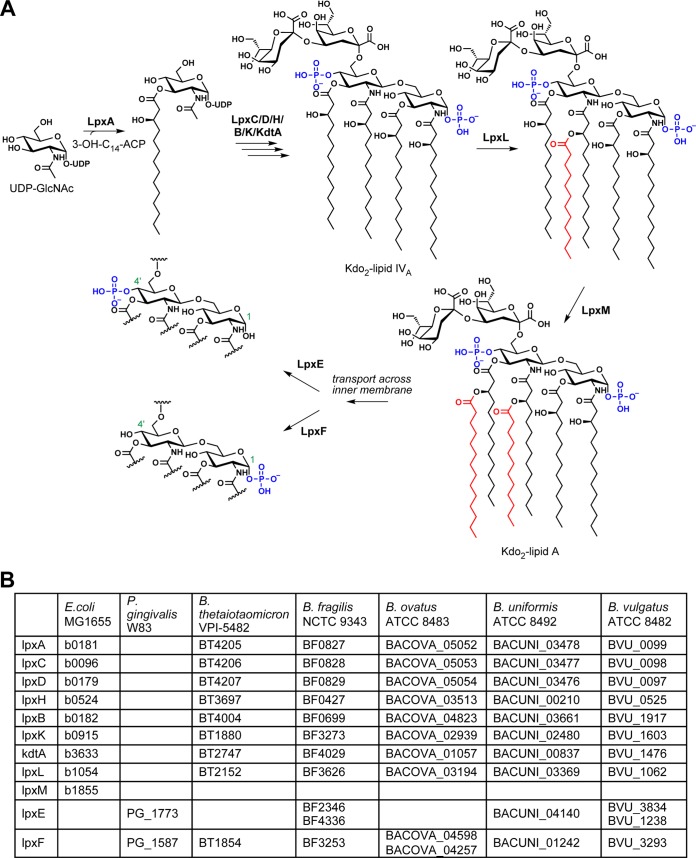
FIG 1 .
The Raetz pathway for lipid A biosynthesis in E. coli and its homologs in Bacteroides. (A) Abbreviated schematic of the Raetz pathway, where starting material UDP-GlcNAc is acylated, glycosylated, and phosphorylated by a series of nine biosynthetic enzymes to produce 3-deoxy-d-manno-octulosonic acid 2 (Kdo2)-lipid A. E. coli produces lipid A that is phosphorylated at both the 1 and 4′ positions on the diglucosamine backbone, but the lipid A 1- and 4′-phosphatases LpxE and LpxF have been identified in other bacteria and are thought to act after biosynthesis of Kdo2-lipid A is complete. (B) Locus tags of homologs of the Raetz pathway genes in E. coli MG1655 and lpxE and lpxF in Porphyromonas gingivalis W83 from a selection of Bacteroides species. Bacteroides has homologs for every gene in the pathway except that it has only one secondary acyltransferase, suggesting that its lipid A is predominantly penta-acylated. The species vary more in their putative homologs of the P. gingivalis phosphatases.