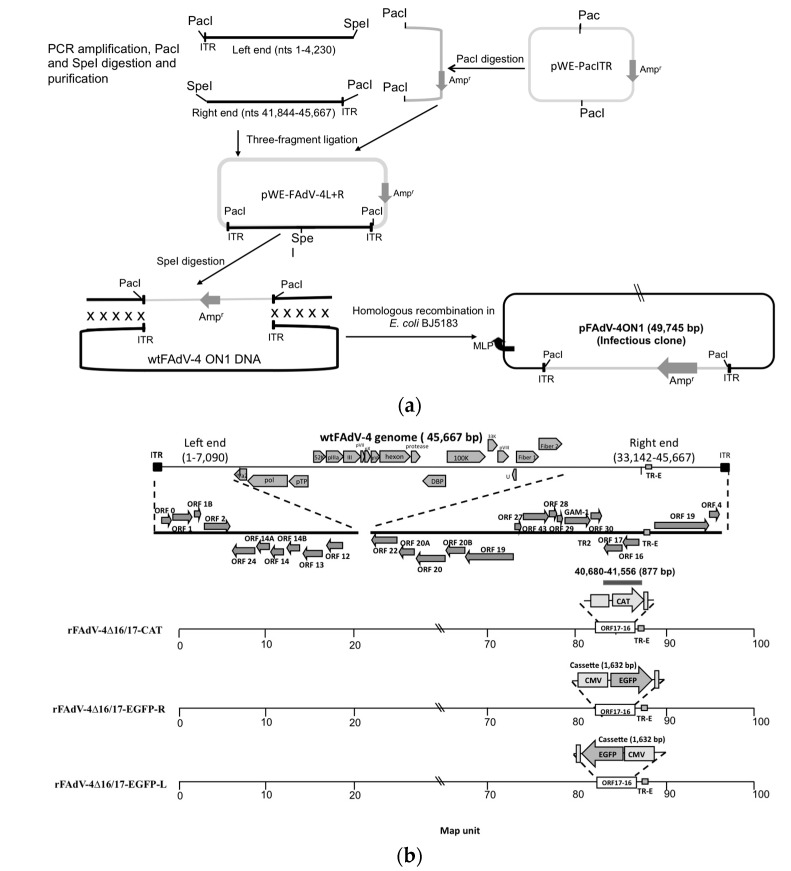
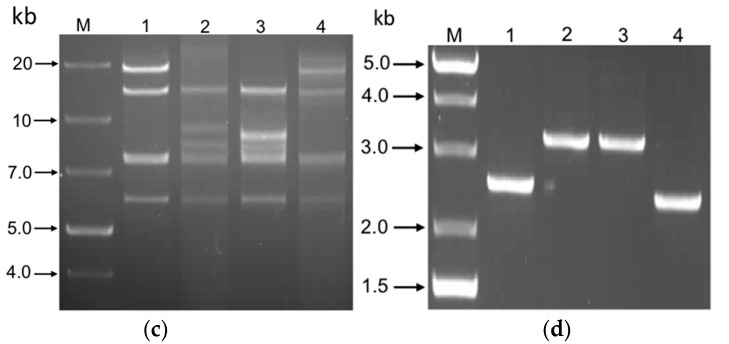
Figure 1.
Construction of fowl adenovirus 4 ON1 (FAdV-4 ON1) infectious clone by homologous recombination and generation of ORF16–17 deleted mutant/recombinant viruses. (a) The left- and right-end termini (1–4230 and 41,844–45,667, respectively) were PCR-amplified with specific primers (Table 1) to introduce PacI and SpeI sites. Polymerase chain reaction (PCR) products were digested with PacI and SpeI and purified as described in materials and methods. The modified pWE-15 cosmid was digested with PacI and purified. The digested PCR fragments and pWE-15 were ligated to generate pWE-FAdV-4L+R, the intermediate construct. pWE-FAdV-4L+R was digested with SpeI, purified, and co-transformed with the FAdV-4 genome in E. coli BJ5183 to generate pFAdV-4 ON1. Viable virus was generated upon transfection of CH-SAH cells with PacI-digested pFAdV-4 ON1 (not shown); (b) chloramphenicol acetyl transferase (CAT)-mediated deletion of ORFs 16 and 17 was carried out using the lambda Red recombinase approach as described previously [21,29] to generate pFAdV-4Δ16/17-CAT. Chloramphenicol acetyl transferase was removed by SwaI digestion and replaced with the enhanced-green fluorescence protein (EGFP) expression cassette (cytomegalovirus promoter-EGFP coding region-Poly A signal) in both leftward and rightward orientations; (c) Infectious clones were verified by HindIII digestion: lane M, 1 kb DNA ladder; lane 1, pFAdV-4Δ16/17-CAT (fragment sizes are 16.3 kb, 12.4 kb, 7.7 kb, 7.4 kb and 5.9 kb); lane 2, pFAdV-4Δ16/17-EGFP-R (fragment sizes are 12.4 kb, 9.4 kb, 7.7 kb, 7.4 kb, 5.9 kb and 5.7 kb); lane 3, pFAdV-4Δ16/17-EGFP-L (fragment sizes are 12.3 kb, 9.0 kb, 7.7 kb, 7.4 kb, 6.0 kb and 5.9 kb); lane 4, parental pFAdV-4 ON1 (fragment sizes are 16.4 kb, 12.4 kb, 7.7 kb, 7.4 kb and 5.9 kb); (d) viable viruses were generated upon transfection of PacI-digested infectious clones (wild type and recombinants). The stability of the transgenes (CAT and EGFP) in viable virus genomes was verified after three passages by PCR with specific primers flanking the deleted region; lane M, 1 kb DNA ladder; lane 1, FAdV-4Δ16/17-CAT (2,628 bp); lane 2, FAdV-4Δ16/17-EGFP-R (3,210 bp); lane 3, FAdV-4Δ16/17-EGFP-L (3,210 bp); and lane 4, wtFAdV-4 ON1 (2,453 bp). ITR: inverted terminal repeat.