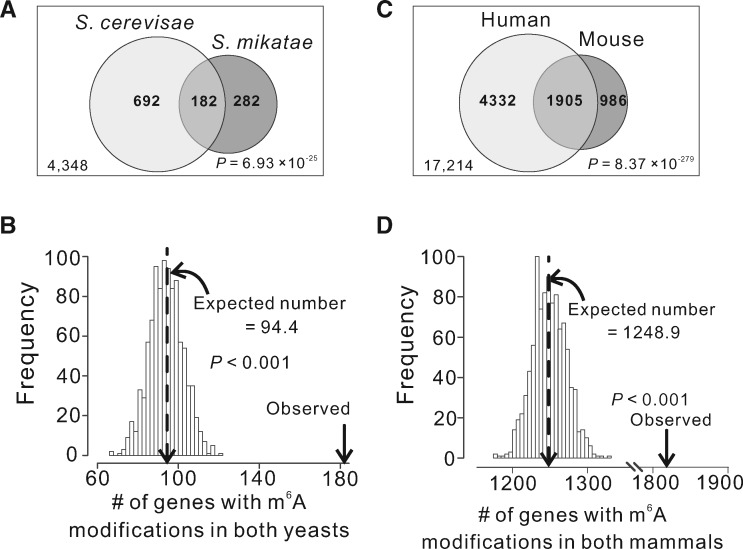
Fig. 3.
Sharing of m6A-modified genes between species. (A) Venn diagram showing the number of one-to-one orthologous genes under comparison (large box, with the number indicated at the lower left corner), as well as the number of m6A-modified genes observed in each yeast species and the number of shared m6A-modified genes. P-value from a hypergeometric test indicates the probability of observation under the null hypothesis of independent m6A modifications of genes in the two yeasts. (B) Frequency distribution of the number of shared m6A-modified genes expected by chance in yeasts upon the control for potential detection bias. The dotted line shows the mean of the distribution, whereas the arrow indicates the number of observed shared m6A-modified genes. P-value is the fraction of the distribution on the right side of the arrow that indicates the observed number. (C) Venn diagram showing the number of one-to-one orthologous genes under comparison (large box, with the number indicated at the lower left corner), as well as the number of m6A-modified genes observed in each mammalian species and the number of shared m6A-modified genes. P-value is from a hypergeometric test. (D) Frequency distribution of the number of shared m6A-modified genes expected by chance in mammals upon the control for potential detection bias. All symbols have the same meanings as in panel B.