Abstract
Field studies were carried out in four Florida counties to investigate winter and spring ecology of host use by Culiseta melanura (Coquillet), the primary vector of eastern equine encephalomyelitis virus (EEEV) in North America. Bloodmeal analysis by PCR was used to identify 233 host bloodmeals, which mainly originated from birds (78.5%) and reptiles (17.2%), primarily Anolis spp. lizards. Across counties, the percentage of bloodmeals from reptiles (7–37% depending upon county) increased with increasing day length and temperature in the spring. Multiple logistic regression revealed that differences in reptile host use across collection sites were largely explained by differences in average day length and temperature on the day of collection, and is probably owing to environment-driven behavioral patterns of ectothermic animals. Although past studies have demonstrated reptile biting by epizootic vectors of EEEV, including Culex (Melanoconion) spp., this is the first study to demonstrate widespread and common feeding upon ectothermic hosts by Cs. melanura. This work suggests that reptiles, particularly anole lizards, play a role in the ecology of EEEV in Florida either as amplifying hosts or as noncompetent hosts which dilute vector feedings thereby suppressing transmission. Detailed laboratory studies investigating impacts of environmental variables (temperature and photoperiod) on EEEV competence of anoles are needed to assess whether these animals support virus amplification.
Keywords: Culiseta, arbovirus, host use, reptile, lizard
Culiseta melanura (Coquillet) is considered the primary enzootic vector of eastern equine encephalomyelitis virus (EEEV), a zoonotic mosquito-borne alphavirus distributed in isolated transmission foci throughout the eastern United States, from Florida to New Hampshire, and extending into the Midwest (Mullen and Durden 2009). Despite high case-fatality rates of the virus in both humans and horses, much remains unknown concerning the ecology of the primary vector throughout parts of the virus’ range, particularly in the southern United States. As a result, EEEV transmission to humans and horses remains cryptic and difficult to predict.
In the northern portion of the EEEV distribution (e.g., Connecticut, Vermont), Cs. melanura adults are active during a relatively short season (June–October; Molaei and Andreadis 2006, Molaei et al. 2015a), which corresponds closely to the EEEV transmission season (Molaei et al. 2015a). Despite relatively long periods each year (8 mo) without apparent EEEV transmission (Armstrong et al. 2008, Molaei et al. 2015a), EEEV phylogenetic studies have concluded that the virus often overwinters locally at northern sites for several years before going locally extinct (Armstrong et al. 2008, Young et al. 2008) based upon closely related strains being recovered during successive years of multiyear epizootics. Interestingly, some EEEV isolates from northern foci (Connecticut, New York) were most genetically similar to EEEV strains from birds in the deep south (Georgia, Florida), suggesting that EEEV may be reintroduced northward by migrating birds that winter in or migrate through the southern United States (Armstrong et al. 2008, Young et al. 2008). In contrast with northern locales, EEEV transmission occurs year-round in Florida (Bigler et al. 1976), and the virus has been isolated from the primary vector, Cs. melanura, throughout the year, including the winter and spring maintenance and amplification periods (Wellings et al. 1972) when Cs. melanura is most abundant (Edman et al. 1972). In Florida, transmission of the virus to humans and horses peaks in June and July (Bigler et al. 1976) when Cs. melanura is relatively uncommon (Edman et al. 1972). Transmission to mammals has often been attributed to secondary vectors, such as Coquillettidia perturbans (Walker) and spring Aedes species in northern locations (Scott and Weaver 1989), Culex erraticus (Dyar and Knab) in Alabama (Bingham et al 2015), and Culex nigripalpus Theobald in Florida (Wellings et al. 1972). Therefore, Cs. melanura ecology during the winter and spring (EEEV maintenance and amplification period) in the southern United States is important both from a local perspective as virus is propagated in birds, but is also potentially important owing to the hypothesized transmission of EEEV to birds migrating northward.
The seasonality of host use is thought to be an important factor driving the timing and intensity of zoonotic mosquito-borne virus epidemics (Kilpatrick et al. 2006). Examples of seasonal shifts in feeding patterns have been recorded in Cx. nigripalpus (Edman and Taylor 1968), a vector of St. Louis encephalitis virus and West Nile virus, Culex tarsalis Coquillett (Tempelis et al. 1965), a vector of western equine encephalitis virus, Culex pipiens L. (Kilpatrick et al. 2006), a vector of West Nile virus, and Cx. erraticus (Burkett-Cadena et al. 2012), a secondary vector of EEEV. In contrast to these Culex species, Cs. melanura has been frequently characterized as highly ornithophilic, taking the vast majority (>90%) of its bloodmeals strictly from avian hosts. However, relatively low percentages of reptilian (Edman et al. 1972, Burkett-Cadena et al. 2008) and mammalian (Edman et al. 1972, Bingham et al. 2014) bloodmeals have been recorded in the few studies conducted in southeastern states. Documenting seasonal trends in Cs. melanura host use may help identify factors affecting early season EEEV amplification in the southern states.
The present study investigated the winter and spring ecology of the primary vector of EEEV, Cs. melanura, focusing on the phenology of host use. Four counties were selected for study, based upon previous evidence of winter and spring seroconversions to EEEV in sentinel chickens (Walton County, Citrus County, and Orange County) or for the ability to compare results with a previous (Edman et al. 1972) bloodmeal study (Indian River County). Eastern equine encephalomyelitis virus transmission is frequently detected in the three northern counties (Walton, Citrus, and Orange), while EEEV transmission is rarely detected in Indian River County (Florida Department of Health). The aims of the research were to investigate patterns of abundance and host utilization of Cs. melanura during the winter and spring (EEEV maintenance and amplification period) in Florida by concurrently sampling from disparate locations within the state. Logistic regression was employed to analyze relationships between seasonal shifts in host use and environmental parameters (temperature and day length) that have been shown to affect the seasonal behaviors of host animals (Bishop and Echternacht 2004).
Materials and Methods
Study Sites
Mosquito collections were conducted in four counties (Walton, Citrus, Orange, and Indian River) which fall within the endemic zone for EEEV transmission, yet vary substantially in their climates and habitats, owing to differences in latitude and proximity to Gulf of Mexico or Atlantic Ocean. All four counties participate in the Florida sentinel chicken arbovirus surveillance program, which has demonstrated historical winter seroconversions to EEEV in sentinel chickens in Walton, Citrus, and Orange counties (Florida Department of Health). Indian River County, where EEEV is less common, has historical data on host utilization by Cs. melanura (Edman et al. 1972). For comparative purposes, localities in this county were also included. Within each county, two to four sites were sampled using six wire-frame resting shelters per site (Burkett-Cadena 2011). In Walton, Citrus, and Orange counties, resting shelters were placed within 500 m of coops at locations where sentinel chicken flocks were operated. In Walton County, two additional sites, “St. Joe” and “County Line,” were added to the study on 2 March 2015 in response to seroconversions to EEEV that were detected in the sentinel chicken flocks at those sites. In Indian River County, three sites were selected in the western portion of the county where Cs. melanura is abundant, although sentinel chicken flocks are not maintained in that (sparsely populated) portion of the county. Schwey hammock, the site of previous research on Cs. melanura host use in Indian River County (Edman et al. 1972), has been developed into orange grove, and the wetland drained, so other sites (<20 km distant) with red maple or cypress swamps were selected.
Mosquito Collections
Mosquito sampling was conducted between December 2014 and May 2015, coinciding with the period of greatest abundance of adult Cs. melanura (Edman et al. 1972). Twice weekly, mosquitoes were aspirated from wire-frame resting shelters using a modified hand-held vacuum (BDH1800S Ni-Cd 18V Dustbuster, Black & Decker, MD) fitted with screen-bottom collection cups (BioQuip Products, Rancho Dominguez, CA). Owing to the large entrance to the resting shelters, methods were used to reduce the numbers of mosquitoes escaping the shelters once disturbed. In Walton, Citrus, and Orange counties, a piece of corrugated plastic cardboard with a hole in the center was placed at the opening of the shelter to create a smaller exit area, diverting exiting mosquitoes to the intake of the aspirator. In Indian River County, the intake of the aspirator was fitted with a funnel that fit snugly into the resting shelter and funneled exiting mosquitoes into the aspirator intake. Once collected, mosquitoes were pooled by site, frozen, and shipped on dry ice to the Florida Medical Entomology Laboratory in Vero Beach for processing. Mosquitoes were identified to species using morphological characteristics (Darsie and Ward 1981), and females of Cs. melanura were pooled by site and date for virus screening. The thoraces of blood-engorged females were manually separated from the abdomen so that bloodmeal analysis could be performed on individual abdomens and pool-screening for EEEV by RT-PCR could be performed on the heads and thoraces of all females (up to 50 per pool).
Bloodmeal Analysis
Host species identification consisted of DNA extraction using Chelex resin (BioRad, Hercules, CA) followed by PCR assays targeting cytochrome b (cytb) and 16s ribosomal RNA (16s) genes and Sanger sequencing. Blood-engorged abdomens of mosquitoes were individually homogenized in 10 µl of 0.9% NaCl solution using a disposable plastic pestle. An addition of 240 µl of preheated (100°C) 20% Chelex resin solution was followed by 10-min incubation in a 100°C water bath. Samples were centrifuged at 6,000 rpm for 5 min, and the supernatant containing the extracted DNA was transferred to a new tube. For each bloodmeal, 2.5 µl of extracted DNA was used as the template in 25-µl PCR amplification reaction. All PCR reactions contained 0.625 µl of each primer (20 µM), 2.5 µl 10× PCR buffer, 1.5 µl MgCl2 (50 mM), 2.5 µl dNTP mix (2 mM each), 0.5 µl Taq DNA polymerase (5 U/µl), and 14.25 µl molecular grade water. PCR products were stained with ethidium bromide, electrophoresed, and visualized on a 1% agarose gel. We used a hierarchical approach to amplify fragments of the vertebrate cytb (avian) or 16s ribosomal RNA (reptile, mammal, and amphibian) genes using four primer pairs, as necessary. As we anticipated the majority of Cs. melanura bloodmeals to be derived from avian hosts, the initial PCR assay performed on all samples used two sets of primers targeting cytb of birds (Lee et al. 2008). The first assay used the primers L0/H1, yielding a 220-bp product primarily used to identify bird bloodmeals (Lee et al. 2008), but which also resulted in amplicons for some nonavian DNA. If no host DNA amplified via either avian-specific PCR, a third PCR assay producing a 450-bp product targeting reptilian DNA was performed using primer pair 16L1/H3056 developed for studies of phylogeography of anoline lizards and xenodontine snakes (Hass et al. 1993, Vidal et al. 2000). If no amplification was achieved using the above protocols, a fourth PCR assay using primers L2513/H2714 of Kitano et al. (2007) that yielded a 200-bp product was performed that targets DNA of mammalian and amphibian hosts (Burkett-Cadena et al. 2008). The Sanger method was used to sequence PCR products of expected size. Sequencing was performed by Eurofins MWG Operon (Huntsville, AL). We used the BLASTn function to compare bloodmeal sequences to referenced sequences on the National Center for Biotechnology and Information database GenBank. Only sequences with ≥95% similarity to referenced GenBank sequences (or reference sequences obtained from tissue samples of known specimens) were considered identified. Sequences derived from tissue samples from road-killed animals (University of Florida IACUC protocol number 201408377) and museum specimens were used to confirm matches to vertebrate host species for which 16s rRNA or cytb sequences were not available in the GenBank database and to verify that PCR protocols effectively amplified DNA of species belonging to a broad range of vertebrate classes and orders (Aves, Mammalia, Squamata, Testudines, and Anura), while excluding amplification of mosquito DNA. Although our hierarchical approach may have missed some mixed-host bloodmeals, previous research (Molaei et al 2006) suggests that Cs. melanura exhibits relatively low rates of mixed host use (5%). In addition, the “avian-specific” primers (L0/H1 and L0/H0) that were used for all samples also produced amplicons for a large proportion of nonavian bloodmeals (53% of reptiles), reducing the likelihood of undetected mixed bloodmeals.
Virus Detection
Mosquito pools were tested for the presence of EEEV as previously described (Bingham et al. 2014). In brief, pools of mosquitoes were homogenized in 1 ml BFD (biological field diluent; 90% minimum essential medium with Hanks’ salts, 10% fetal bovine serum, 200 U/ml penicillin, 200 mg/ml streptomycin, 2.5 mg/ml amphotericin B, and 50 mg/ml kanamycin) using TissueLyser II (QIAGEN, Valencia, CA) for 4 min at 25 Hz. The resulting homogenate was centrifuged at 10,000 rpm for 3 min at 4°C. Viral RNA was isolated from 140 µl of the homogenate supernatant using QIAamp viral RNA kit (QIAGEN, Valencia, CA), according to the manufacturer’s protocol. A real time reverse transcriptase-polymerase chain reaction (qRT-PCR) assay was conducted using the iScript one step RT-PCR kit for probes (Bio-Rad, Hercules, CA), according to the manufacturer’s protocol. The primers, probe, and reaction conditions used to detect EEEV RNA were those recommended by Lambert and others (2003), with the exception that reactions were performed in a final volume of 25 µl, and used 5 µl of the RNA template. Samples producing a signal at a Ct value of 37 or below were considered putatively positive. RNA samples found to be putatively positive in the initial assay were subjected to a confirmatory qRT-PCR assay as previously described (Lambert et al. 2003). Samples producing a positive signal with a Ct value of <40 in the confirmatory assay were scored as confirmed positive. A ∼200-bp fragment generated from cloned amplicons of EEEV cDNA was used as a positive control for EEEV detection.
Data Analysis
The chi-square test of independence was used to test for differences in the monthly distributions of major wild host groups (excluding chickens), aggregated across sites. Chickens were excluded from this analysis because they were not present at one quarter of total sampling sites.
Multiple logistic regression analysis was used to model the probability that individual Cs. melanura bloodmeals originated from reptilian hosts using nightly low temperature, day length, and county as predictors (PROC LOGISTIC, SAS Institute). Nightly low temperature was included in the model as it is a measure of overnight cooling, a likely determinant of ectothermic host availability during the host-seeking period of Cs. melanura. Day length was included as a predictor of reptile seasonal availability, as activity patterns and biological processes such as feeding, growth, and onset of reproduction are stimulated by lengthening spring days in many taxa (Bradshaw and Holzapfel 2007). The response variable “host class” was coded as either “reptile” or “other”. Data from Orange County were excluded from the analysis owing to small total sample size (n = 6). The analyzed model set contained all submodels and interaction combinations with the predictors’ day length, temperature, and county, and models were compared using AICc. Temperature data were obtained from the National Climatic Data Center weather stations from Fort Walton Beach Airport in Destin (Walton County), Inverness (Citrus County), Orlando West (Orange County), and Vero Beach (Indian River County; http://www.ncdc.noaa.gov, accessed 27 October 2016). Weather stations containing the most complete data set and located closest to the sampling sites (range 7–45 km) were selected for use in the analysis. Day length data (minutes of daylight from sunrise to sunset) were obtained using The United States Naval Observatory, Sun or Moon Rise/Set Table tool found at http://www.usno.navy.mil/USNO/, last accessed 27 October 2016.
Multiple logistic regression analysis was used to model the probability that Cs. melanura bloodmeals from wild bird species were taken from nonresident (migratory) host species using the same predictors as above (day length and county). Models were compared by AICC using data coded as either “resident” or “nonresident”. Because sample size was small for Orange County, this analysis was limited to bloodmeals taken from wild birds in Walton, Citrus, and Indian River counties.
Results
Mosquito Collections
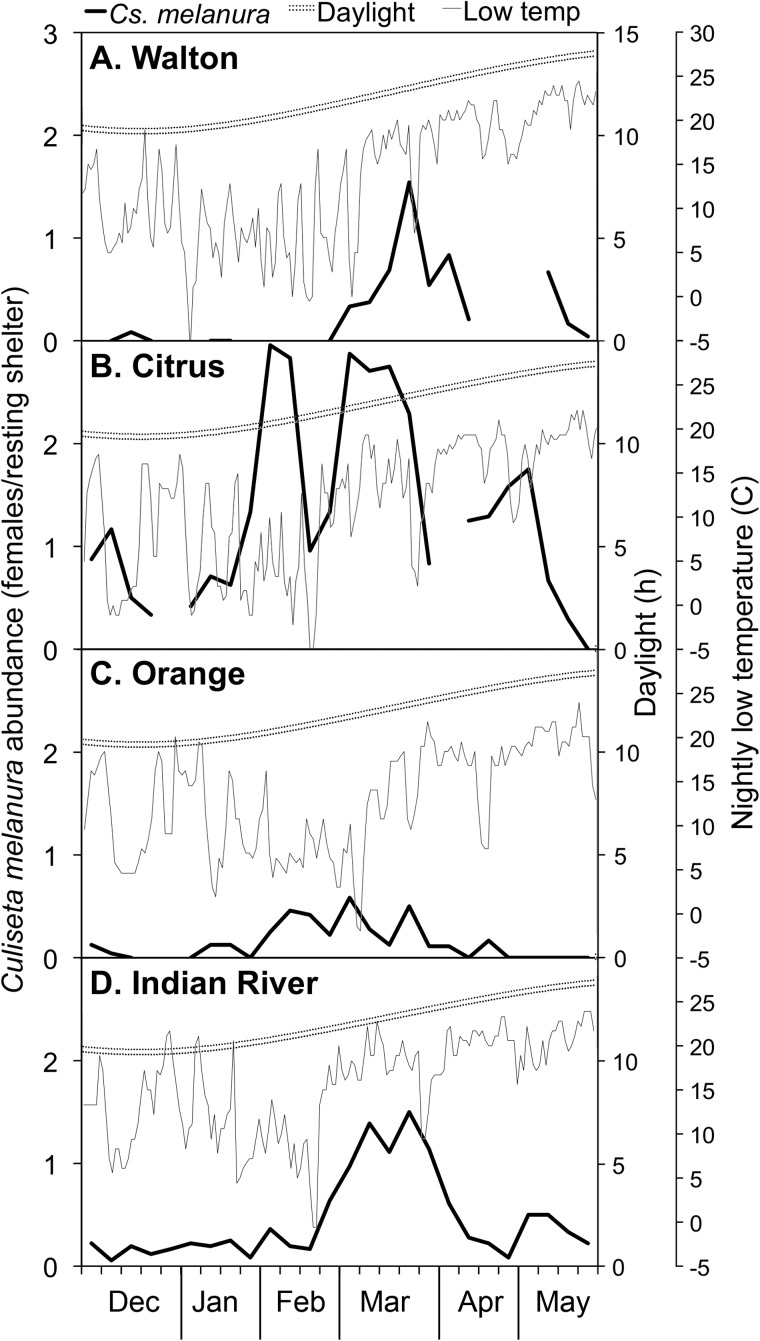
Across all four counties, a total of 1,298 Cs. melanura females, including 254 blood-fed individuals, were collected during the sampling period. Overall, very few Cs. melanura females were captured during the period from December to January (Fig. 1) compared with the period of February to May. Culiseta melanura was most abundant in March and April, 2015 (Fig. 1), in Indian River and Walton counties, with numbers peaking in late March (1.50 females per shelter in Indian River and 1.54 females in Walton). In Citrus County, two peaks in Cs. melanura populations were observed: one that coincided with the March abundance peaks in Indian River and Walton counties, and another in February (Fig. 1). An additional, smaller late spring peak was observed in May in all three of these counties.
Fig. 1.
Average number of female Cs. melanura captured per shelter each week in four Florida counties between December 2014 and May 2015. Day length and nightly low temperature are given for each county. Gaps in data represent periods during which resting shelters were not sampled. Error estimates cannot be calculated, as mosquitoes were pooled by date from each trapping site before identification.
Bloodmeal Analysis
Vertebrate hosts were identified to the species level for 233 (91.7%) of 254 blood-engorged Cs. melanura females (Table 1). Avian species composed the majority of overall hosts (78.5% of total), although the percentage of bloodmeals derived from birds varied by county (Fig. 2), from just 56.7% avian in Walton County to 100.0% avian in Orange County (Fig. 2; Table 1). Chickens were the most common (Citrus and Orange counties) or second most common (Walton County) host species in counties in which sampling points were located near sentinel chicken flocks. Among bloodmeals from wild birds, two year-round resident species, northern cardinal, Cardinalis cardinalis (16.3% of all bloodmeals), and white-eyed vireo, Vireo griseus (8.6% of all bloodmeals), were dominant in Citrus and Indian River counties. Northern cardinal was the only host detected in all four counties. Two nonresident species, eastern phoebe, Sayornis phoebe, and common yellowthroat, Geothlypis trichas, each comprised 8.6% of bloodmeals from Indian River County and 3–4% of total bloodmeals. Other notable (>1%) nonresident species include hermit thrush, Catharus guttatus (2.6% of total), yellow-rumped warbler, Setophaga coronata (2.2%), American robin, Turdus migratorius (1.7%), and ruby-crowned kinglet, Regulus calendula (1.7%). Green heron, Butorides virescens, was the only wading bird species commonly fed upon, constituting 3.0% of total bloodmeals. Golden-crowned warbler, Basileuterus culicivorus, a species not previously recorded from the United States, was identified with 95–97% sequence similarity to referenced sequences (2.2% of total bloodmeals) and likely represents an avian host species for which published sequences are not currently available.
Table 1.
Bloodmeal hosts of Cs. melanura from four Florida counties, November 2014 to May 2015
| Common name | Scientific name | RCa | Citrus | Indian River | Orange | Walton | Total (%) |
|---|---|---|---|---|---|---|---|
| Avian | 92 (88.5) | 68 (73.1) | 6 (100.0) | 17 (56.7) | 183 (78.5) | ||
| Chicken | Gallus gallus | E | 32 (30.7) | 3 (50.0) | 5 (16.7) | 40 (17.2) | |
| Northern cardinal | Cardinalis cardinalis | R | 25 (24.0) | 9 (9.7) | 1 (16.7) | 3 (10.0) | 38 (16.3) |
| White-eyed vireo | Vireo griseus | R | 5 (4.8) | 15 (16.1) | 20 (8.6) | ||
| Eastern phoebe | Sayornis phoebe | N | 1 (1.0) | 8 (8.6) | 9 (3.9) | ||
| Common yellowthroat | Geothlypis trichas | N | 8 (8.6) | 8 (3.4) | |||
| Green heron | Butorides virescens | R | 5 (5.4) | 2 (6.7) | 7 (3.0) | ||
| Hermit thrush | Catharus guttatus | N | 4 (3.8) | 2 (6.7) | 6 (2.6) | ||
| Yellow-rumped warbler | Setophaga coronata | N | 2 (1.9) | 2 (2.2) | 1 (3.3) | 5 (2.2) | |
| Golden-crowned warblerb | Basileuterus culicivorus | E | 3 (2.9) | 2 (2.2) | 5 (2.2) | ||
| American robin | Turdus migratorius | N | 4 (3.8) | 4 (1.7) | |||
| Ruby-crowned kinglet | Regulus calendula | N | 2 (1.9) | 2 (6.7) | 4 (1.7) | ||
| Blue Jay | Cyanocitta cristata | R | 1 (1.0) | 3 (3.2) | 4 (1.7) | ||
| Barred owl | Strix varia | R | 1 (1.0) | 2 (2.2) | 3 (1.3) | ||
| Red-winged blackbird | Agelaius phoeniceus | R | 3 (3.2) | 3 (1.3) | |||
| Carolina wren | Thryothorus ludovicianus | R | 2 (1.9) | 1 (3.3) | 3 (1.3) | ||
| Palm warbler | Setophaga palmarum | N | 1 (1.0) | 1 (1.1) | 2 (0.9) | ||
| Blue-headed vireo | Vireo solitarius | N | 1 (1.0) | 1 (1.1) | 2 (0.9) | ||
| American crow | Corvus brachyrhynchos | R | 2 (1.9) | 2 (0.9) | |||
| Blue-gray gnatcatcher | Polioptila caerulea | R | 1 (1.0) | 1 (1.1) | 2 (0.9) | ||
| Barn owl | Tyto alba | R | 1 (1.1) | 1 (0.4) | |||
| Yellow-throated Vireo | Vireo flavifrons | N | 1 (1.0) | 1 (0.4) | |||
| White-tipped doveb | Leptotila verreauxi | E | 1 (1.1) | 1 (0.4) | |||
| American bittern | Botaurus lentiginosus | N | 1 (1.1) | 1 (0.4) | |||
| Red-eyed vireo | Vireo olivaceus | N | 1 (3.3) | 1 (0.4) | |||
| Common ground dove | Columbina passerina | R | 1 (16.7) | 1 (0.4) | |||
| Chipping sparrow | Spizella passerina | N | 1 (16.7) | 1 (0.4) | |||
| Eastern Whip-poor-will | Caprimulgus vociferus | N | 1 (1.0) | 1 (0.4) | |||
| Wood thrush | Hylocichla mustelina | N | 1 (1.0) | 1 (0.4) | |||
| Red crossbillb | Loxia curvirostra | E | 1 (1.1) | 1 (0.4) | |||
| Pine warbler | Setophaga pinus | N | 1 (1.0) | 1 (0.4) | |||
| Cedar waxwing | Bombycilla cedrorum | N | 1 (1.0) | 1 (0.4) | |||
| Black-crowned night heron | Nycticorax nycticorax | R | 1 (1.1) | 1 (0.4) | |||
| Limpkin | Aramus guarauna | R | 1 (1.1) | 1 (0.4) | |||
| Marsh wren | Cistothorus palustris | N | 1 (1.1) | 1 (0.4) | |||
| Gray catbird | Dumetella carolinensis | N | 1 (1.1) | 1 (0.4) | |||
| Mammalian | 5 (4.8) | 3 (3.2) | 0 | 2 (6.7) | 10 (4.3) | ||
| Human | Homo sapiens | 4 (3.8) | 3 (3.2) | 2 (6.7) | 9 (3.9) | ||
| Southern flying squirrel | Glaucomys volans | 1 (1.0) | 1 (0.4) | ||||
| Reptilian | 7 (6.7) | 22 (23.7) | 0 | 11 (36.7) | 40 (17.2) | ||
| Green anole | Anolis carolinensis | 5 (4.8) | 15 (16.1) | 10 (33.3) | 30 (12.9) | ||
| Brown anole | Anolis sagrei | 1 (1.0) | 5 (5.4) | 6 (2.6) | |||
| Banded water snake | Nerodia fasciata | 1 (1.0) | 2 (2.2) | 1 (3.3) | 4 (1.7) | ||
| Total | 104 | 93 | 6 | 30 | 233 |
Numbers in parentheses represent the percentage of column total.
Residency Codes: R—year-round resident; N—nonresident migrant; E—exotic species nonnative to Florida.
Exotic avian host matches may represent native bird species lacking sequences in GenBank.
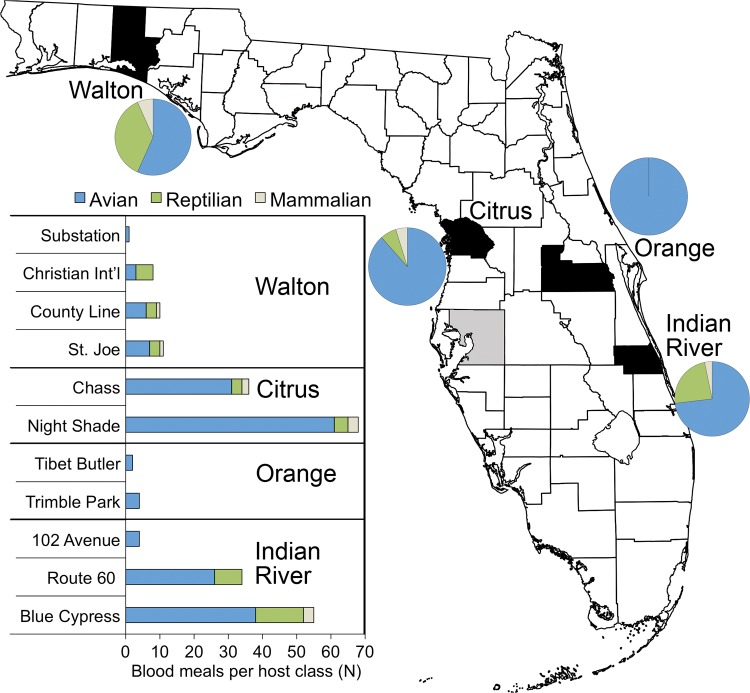
Fig. 2.
Map of Florida counties and class-level host use by Cs. melanura. Counties in which mosquitoes were sampled during this study are shaded black. Hillsborough County, site of Bingham et al. (2014), is shaded gray. Pie charts represent total Cs. melanura bloodmeals aggregated from two to four sites, across 6 mo of field sampling (December 2014–May 2015). Bar charts represent raw numbers from each site within counties.
Reptilian hosts accounted for 17.2% of the total Cs. melanura bloodmeals (Table 1), although the percentage of bloodmeals derived from reptiles varied by county, from relatively low (0% in Orange County and 6.7% in Citrus County; Fig. 2) to relatively high (23.7% in Indian River County and 36.7% in Walton County; Fig. 2). Reptile-derived bloodmeals consisted entirely of two lizard species (Anolis carolinensis, green anole, 12.9%, and Anolis sagrei, brown anole, 2.6%) and one snake species (Nerodia fasciata, banded water snake, 1.7%). Overall, the green anole was the third most frequently fed upon host, following chicken and northern cardinal, but was the most commonly fed upon host in both Walton County and Indian River County (tied with white-eyed vireo). At every site where at least five Cs. melanura bloodmeals were identified, multiple bloodmeals derived from reptilian hosts were observed (Fig. 2). Among these sites, the percentage of reptilian-derived bloodmeals ranged from 5.9% (Nightshade) to 62.5% (Christian International). Mammalian hosts consisted of humans (3.9%) and one southern flying squirrel, together comprising 4.3% of total bloodmeals.
Seasonality of Host Use
Host use by Cs. melanura varied seasonally (Fig. 3) with respect to major host groups (wild birds, mammals, and reptiles; χ2 = 40.6; df = 10; P < 0.001). Birds were the most commonly fed upon class of wild hosts during the majority of sampling months (Fig. 3); however, the percentage of bloodmeals from wild birds was much higher from December to March (76.5–87.5% of wild animal bloodmeals) than April to May (35.3–55.2%). Use of reptilian hosts was strongly seasonal, with 97.5% of reptilian bloodmeals detected from March to May. The paucity of bloodmeals attributed to reptiles in the preceding period (December–February) was not owing to low sample size, as 28.8% of total wild animal bloodmeals were collected in that period. Excluding one early outlier (Fig. 4), reptilian host use began near synchronously at widespread sites in early March, as first bloodmeals from reptiles were detected from samples in Walton (4 March), Citrus (5 March), and Indian River (6 March) counties (Fig. 3).
Fig. 3.
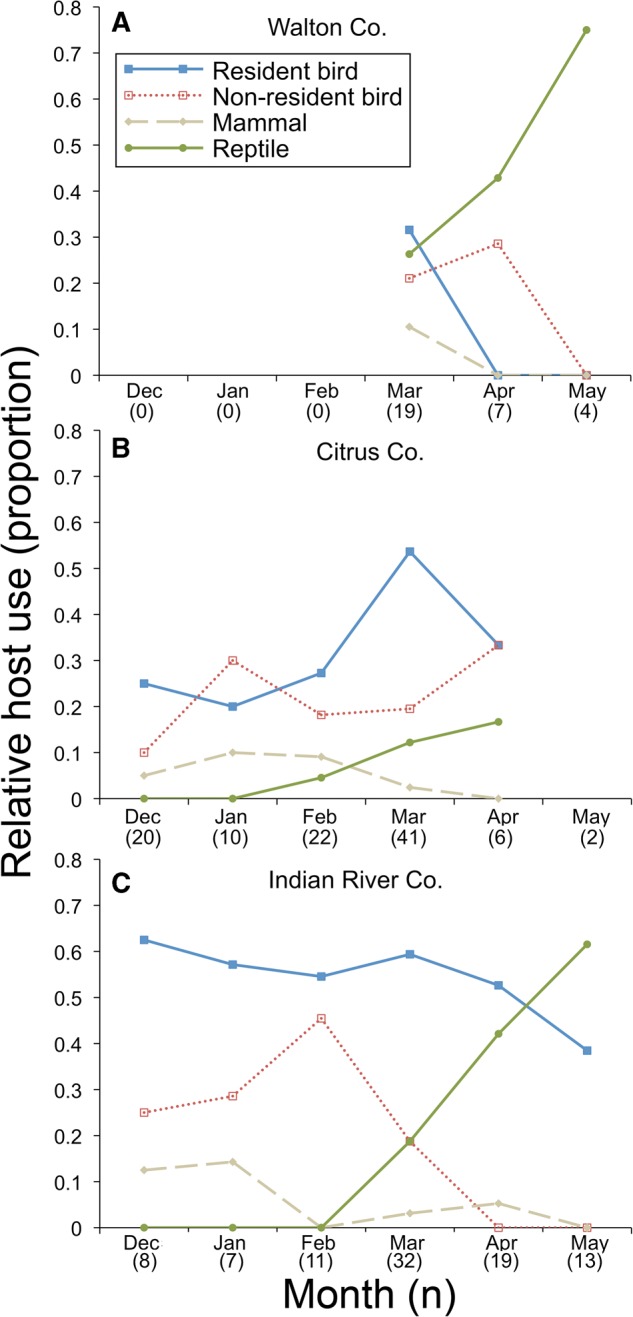
(A–C) Proportions of Cs. melanura bloodmeals from host groups (resident bird, nonresident bird, mammal, and reptile) during the period of December 2014 through May 2015 in (A) Walton County, (B) Citrus County, and (C) Indian River County, FL, United States. Data for chickens, exotic birds, or months in which fewer than four bloodmeals were analyzed are not presented. No blood-engorged Cs. melanura were collected in December through February in Walton County. Samples sizes in parentheses.
Fig. 4.
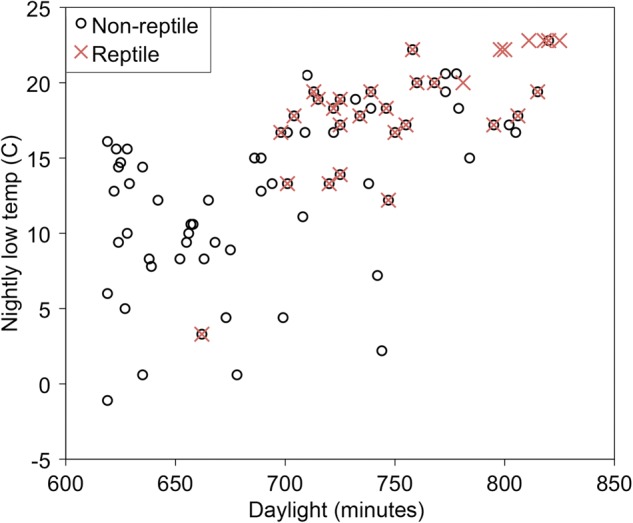
Relationship between day length, nightly low temperature, and host use by Cs. melanura. Points represent bloodmeals collected on a given day from reptile (red X) and nonreptile (open circle) hosts sampled at various day length and temperature combinations. Data are from field samples, aggregated from four Florida counties, December to May (2014–2015). Multiple bloodmeals (identical temperature and day length) are represented by a single point for 58 observations.
The logistic regression model that included the variables “temperature” and “day length” together best explained the probability that Cs. melanura bloodmeals originated from reptiles (Table 2). This combined model fit the response data considerably better than models containing the day length and temperature predictors individually (evidence ratios indicate the combined model has 17.3 and 27.3 times the weight of evidence as the individual models). Three additional models fell within 4 ΔAICC of the top model, indicating substantial support from the data. These models contained the top model with added predictors, including a temperature interaction with day length or effects of county and county × day length interaction (Table 2). Despite increased explanatory power, these more complex models did not overcome the AIC penalty for added parameters. However, a larger sample size could increase support for more complex models. Though models containing “county” increased explanatory power, the absence of this predictor in the top model indicates that differences between counties in the percentage of bloodmeals taken from reptiles (Fig. 2) can be largely explained by differences in day length and temperature on the day of collection. Considering the entire dataset, no reptilian-derived bloodmeals were detected when the nightly low temperature was <12°C or day length was <11.5 h (690 min; Fig. 4), excepting one outlier (green anole, 3°C and 11-h day length on 9 February, Citrus County). At higher temperature and day length values, the proportion of reptilian-derived bloodmeals increased logistically with increasing day length and temperature (Fig. 3). The top model (day length and temperature) estimated parameter values are −15.0634 (95% CI: −22.0815, −8.7909) for the intercept, 0.0148 (95% CI: 0.00484, 0.0253) for day length, and 0.17 (95% CI: 0.0464, 0.3176) for temperature. Nagelkerkle’s R-squared () value (Nagelkerke 1991) of 0.322 indicates the predictive power of the model.
Table 2.
Comparison of logistic regression models predicting the probability that bloodmeals of Cs. melanura originated from reptile hosts
| Parametersa | n | AICC | Δ AICC | wi | ERb | |
|---|---|---|---|---|---|---|
| T, D | 227 | 168.24 | 0.00 | 0.41 | 1.00 | 0.322 |
| T, D, T × D | 227 | 169.77 | 1.53 | 0.19 | 2.15 | 0.330 |
| T, D, C | 227 | 170.92 | 2.67 | 0.11 | 3.81 | 0.325 |
| T, D, C, D × C | 227 | 171.61 | 3.37 | 0.08 | 5.38 | 0.350 |
| T, D, C, T × D | 227 | 172.56 | 4.32 | 0.05 | 8.67 | 0.333 |
| D, C | 227 | 172.88 | 4.64 | 0.04 | 10.16 | 0.306 |
| T, D, C, T × D, D × C | 227 | 173.46 | 5.22 | 0.03 | 13.61 | 0.351 |
| D | 227 | 173.94 | 5.70 | 0.02 | 17.25 | 0.276 |
| T, D, C, T × C | 227 | 174.35 | 6.11 | 0.02 | 21.25 | 0.334 |
The analysis includes all bloodmeal data from Walton, Citrus, and Indian River Counties combined. All submodels and interactions containing the predictors’ day length, temperature, and county were compared, and the top 10 models are presented in the table.
Parameters include nightly low temperature (T), day length (D), and county (C) of collected bloodmeal.
The weight of evidence, wi, is the probability that the given model best fits the data with the sum of all model probabilities equal to one. The evidence ratio, ER, is the likelihood of the best model compared with the current model.
Analysis of all data on resident versus nonresident bird host use indicated that the best model included “day length” and “county” as predictors (Supp. Table 1 [online only]). However, the best model was within 1.02 ΔAICC of the intercept-only model and showed very low predictive power ( = 0.074), indicating that the chosen predictors were not useful for explaining the data.
Virus Detection
None of the 50 pools of female Cs. melanura, representing 1,298 individuals, tested positive for EEEV by RT-PCR.
Discussion
Females of Cs. melanura displayed a shift toward increased feeding upon reptiles in spring months, which was driven by a combination of increasing day length and temperature (Fig. 4). The county of origin had a limited effect on this pattern, indicating that this is a broadly occurring pattern. Furthermore, differences in percentages of reptilian-derived bloodmeals between counties (Fig. 2) can be largely explained by differences in average day length and temperature when samples were collected (Table 2; Fig. 4). Although the model’s R-square value ( =0.32) demonstrates the existence of additional, unknown factors affecting bloodmeal source, the pseudo R-square values in logistic regression are normally low compared with the R-square values of linear regression, and the value of the current model indicates that the included predictors are strong (Hosmer et al. 2013). Both day length and temperature are known to strongly affect the behavior of diverse animals (Bradshaw and Holzapfel 2007), and, in ectotherms, photoperiod and temperature control phenological patterns of activity, growth, and reproduction. The primary mechanism by which day length and temperature affect seasonal patterns of host use by Cs. melanura is probably through effects on the availability of reptilian hosts. Reptiles in the southern United States undergo varying levels of brumation (winter inactivity), spending winter nights in refuges such as rock crevices (Bishop and Echternacht 2004) or under tree bark (Neill 1948), a behavior which reduces their availability to host-seeking mosquitoes. The cavity-roosting behavior of woodpeckers has been invoked as a similar explanation for why these birds are seemingly avoided by Cs. melanura (Edman et al. 1972). As day length and temperature increase, and more reptiles exit brumation, their availability to host-seeking Cs. melanura increases, and a corresponding greater proportion of bloodmeals are taken from these animals. At equivalent day lengths, reptiles were fed upon when temperatures were higher (Fig. 4).
Despite the varying latitudes of sampling sites (2.5° between northernmost and southernmost sites), few clear differences in seasonal patterns of host use were detected between counties in the present study. For example, the spring initiation of reptile biting occurred nearly concurrently in all three counties analyzed, with the first recorded reptilian-derived bloodmeals occurring within a 3-d period (4–6 March). Seasonal changes in the occurrence of bloodmeals from nonresident birds were not detected using predictors of day length and temperature. Because various nonresident birds are known to occur in Florida through late May, a stronger seasonal effect on the probability of observing bloodmeals derived from nonresident birds would likely result from continued sampling into the summer months. However, as Edman et al. (1972) note, Cs. melanura is most common from fall through spring in Florida and Cs. melanura populations in all three counties had reached low levels by the end of May (Fig. 1), decreasing the likelihood that a useful sample size could be collected. Despite the inability to detect seasonal changes in nonresident bird use, several nonresident species (eastern phoebe, yellow-rumped warbler, and hermit thrush), which each represented 2–4% of total bloodmeals in the current work, were also hosts of Cs. melanura from previous studies in Florida (Bingham et al. 2014, Burkett-Cadena et al. 2015). Additionally, relatively infrequent feeding occurred on the American robin (1.7%) and wood thrush (0.4%), two nonresident species that are frequently found to be the most fed-upon hosts of Cs. melanura at northern EEEV foci (Molaei et al. 2006, 2015a, 2016). These species may be of interest in future investigations of virus transport from southern to northern foci.
Although a predominance of bloodmeals derived from birds was expected, given the results of previous research, a large and unexpected percentage of Cs. melanura bloodmeals (17.2%) were derived from reptiles (Anolis spp. lizards and banded water snake). Reptilian-derived bloodmeals were detected at all collection sites where five or more blood-engorged females of Cs. melanura were collected, suggesting that reptile biting by Cs. melanura is widespread in Florida (Fig. 2). Other studies investigating host use by Cs. melanura have generally reported much lower rates (0–2%) of reptile biting by this mosquito (Edman et al. 1972, Bingham et al. 2014), warranting further discussion. The classic study by Edman et al. (1972) utilizing antibody-based bloodmeal analysis (precipitin tests) reported <2% of Cs. melanura bloodmeals originated from reptiles (three snakes, one lizard, four unknown) at sites in Vero Beach and Tampa, FL. A subsequent report (Edman 1979) indicated that “lizard antisera (produced with Iguana serum) reacted poorly with the blood of local (Anolis) lizards” and that, for Culex pilosus (Dyar and Knab), “many of the negative, discarded bloodmeals were likely of lizard origin,” as the creation of a new, broadly reactive lizard antiserum revealed high rates of lizard-biting in Cx. pilosus (Edman 1979). This technical difficulty may also have influenced the number of reptile bloodmeals detected in Edman’s prior study of Cs. melanura (Edman et al. 1972). However, as the number of unidentifiable bloodmeals was not reported, this explanation is speculative.
The low frequency of reptile biting observed in more recent Florida studies utilizing PCR-based methods of bloodmeal analysis may be owing to the timing of the mosquito collections. Samples of blood-engorged Cs. melanura in Bingham et al. (2014) were collected primarily during January and February, which corresponds to a period of very low (2%) reptilian host use in the current work (Fig. 3). Identical resting shelters were used in Bingham et al. (2014) and the current study, so it is unlikely that field methods contributed to the differences in reptile biting observed between the two studies. However, anoles were frequently observed in close proximity to, or even within resting shelters during daytime collecting forays. This suggests the possibility that the use of resting shelters, the dominant method for collecting engorged Cs. melanura in nearly all studies, may have influenced the number of observed anole-derived bloodmeals in our results. Specimens of Cs. melanura (n = 2) aspirated directly from sleeping anoles (Fig. 5) at locations without resting shelters demonstrate that Cs. melanura does feed upon reptiles in nature.
Fig. 5.

Female of Cs. melanura biting a sleeping lizard (Anolis carolinensis) in nature (Columbia County, FL). The mosquito was aspirated, identified, and bloodmeal origin confirmed by PCR. Photo by L. Reeves.
The role of reptiles in transmission of EEEV is dependent on their reservoir competence and vector contact rates. Anoles were the most frequently fed upon reptiles in the current study, satisfying the latter of the above criteria. Previous laboratory infection studies found that most individual anoles produced circulating viremia, although at titers lower than those of snakes (White et al. 2011), and below titers (104 PFU/ml) that are likely to infect mosquitoes. Although few data exist pertaining to EEEV infection in wild Anolis spp. lizards, teiid and iguanid lizards were frequently seropositive for South American strains of EEEV in Panama (Craighead et al. 1962) and EEEV was isolated from a rock iguana in Cuba (Berezin 1977). This suggests that lizards in other regions are regularly exposed to the virus and some even support replication of EEEV. Additionally, in the current study, water snakes constituted 1.0–3.3% of bloodmeals, by county. Although the reservoir competence of water snakes for EEEV has not been evaluated in the lab, garter snakes produced high EEEV titers in laboratory studies (White et al. 2011) and wild cottonmouth snakes had circulating EEEV RNA and antibodies (Graham et al. 2012, Bingham et al. 2012).
Given the relatively high extent of anole biting observed in the current study, anole host use may either enhance or suppress EEEV transmission, depending on reservoir competence of these animals. In either case, the timing of Cs. melanura spring emergence is important as a strong determinant of the proportion of bloodmeals coming from anoles (Figs. 1 and 4). If the environmental factors controlling Cs. melanura emergence can vary independently of the factors controlling reptile availability as hosts, then the degree of synchrony on a given year may influence the extent of EEEV amplification. Previous studies from northern locations have found that heat summation models tracking water temperature of larval habitat strongly predict the timing of Cs. melanura spring emergence (Mahmood and Crans 1998, Andreadis et al. 2012). However, the county-level differences in the seasonality of mosquito abundance in the current study (Fig. 1) resulting in differential host use (Fig. 2) are not easily explained and deserve further study.
Although evidence has been building that reptiles may be involved in an EEEV cycle in the southeastern United States, little evidence supports similar involvement in the northern states. An early serosurvey of ectotherms in Massachusetts found only one EEEV-seropositive turtle of 190 reptiles examined (Hayes et al. 1964), and studies using modern PCR bloodmeal analysis have found little-to-no evidence for reptile biting by Cs. melanura (Molaei et al. 2015a,b). These studies suggest that reptiles do not play a substantial role in EEEV maintenance in northern locales and the ecology of seasonal transmission is substantially different between northern and southern foci, but that disjunct foci may be linked though migratory birds.
In conclusion, field studies investigating seasonal patterns of host use by Cs. melanura in Florida revealed a shift from feeding almost exclusively upon birds during the coldest winter months (shorter days, lower temperatures) to increased feeding upon reptiles (mostly lizards) with longer spring day lengths and higher temperatures. The shift toward feeding upon reptiles is likely owing to increasing availability of these animals during the host-seeking period, as the ectothermic animals become more likely to sleep on exposed surfaces as nightly temperature increases in spring. That the increase in reptile biting occurs nearly synchronously with a spring peak in Cs. melanura abundance suggests that reptiles influence transmission of EEEV in Florida, either through enhancing amplification (as competent hosts) or acting as a sink and suppressing transmission (as dilution hosts). Factors affecting the timing of Cs. melanura spring emergence deserve further investigation, as timing of emergence can have a major influence on which host classes are fed upon. The possible role of Anolis spp. lizards and water snakes in EEEV ecology also warrants further study.
Acknowledgments
We thank Mark Kartzinel and Michael DeBlasio for assisting in field collections in Indian River County. Numerous mosquito control personnel performed field sampling or facilitated the project in Orange County (Tom Breaud, Amador Rodriguez, Bill Casteel, and Sam LeBron), Citrus County (Joel Jacobson, Don Cruea, George Deskins, and Bill Kellner) and Walton County (Ben Brewer). Edzard van Santen, IFAS Statistical Consulting Unit, provided valuable statistical consultations. Donna Dittman and LSU Museum of Natural Science Collection of Genetic Resources provided tissue loans of Vireo griseus (B-71156, B-71157) used to produce representative sequences. An earlier draft was substantially improved based upon comments from two anonymous referees. This work was supported by National Institutes of Health (Grant R56AI01372), Florida Department of Agriculture and Consumer Services project 021805, and NIFA FLA-VME-005446.
References Cited
- Andreadis T. G., Shepard J. J., Thomas M. C.. 2012. Field observations on the overwintering ecology of Culiseta melanura in the northeastern USA. J. Am. Mosq. Control Assoc. 28: 286–291. [DOI] [PubMed] [Google Scholar]
- Armstrong P. M., Andreadis T. G., Anderson J. F., Stull J. W., Mores C. N.. 2008. Tracking eastern equine encephalitis virus perpetuation in the northeastern United States by phylogenetic analysis. Am. J. Trop. Med. Hyg. 79: 291–296. [PubMed] [Google Scholar]
- Berezin V. V. 1977. Characteristics of the ecology of the eastern equine encephalomyelitis virus in the Republic of Cuba. Voprosy Virusologii 1: 62–70. [PubMed] [Google Scholar]
- Bigler W. J., Lassing E. B., Buff E. E., Prather E. C., Beck E. C., Hoff G. L.. 1976. Endemic eastern equine encephalomyelitis in Florida: A twenty-year analysis, 1955–1974. Am. J. Trop. Med. Hyg. 25: 884–890. [DOI] [PubMed] [Google Scholar]
- Bingham A. M., Graham S. P., Burkett-Cadena N. D., White G. S., Hassan H. K., Unnasch T. R.. 2012. Detection of eastern equine encephalomyelitis virus RNA in North American snakes. Am. J. Trop. Med. Hyg. 87: 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. M., Burkett-Cadena N. D., Hassan H. K., McClure C.J.W., Unnasch T. R.. 2014. Field investigations of winter transmission of eastern equine encephalitis virus in Florida. Am. J. Trop. Med. Hyg. 91: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. M., Burkett-Cadena N. D., Hassan H. K., Unnasch T. R.. 2015. Vector competence and capacity of Culex erraticus (Diptera: Culicidae) for eastern equine encephalitis virus in the Southeastern United States. J. Med. Entomol. 53: 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. C., Echternacht A. C.. 2004. Emergence behavior and movements of winter-aggregated green anoles (Anolis carolinensis) and the thermal characteristics of their crevices in Tennessee. Herpetologica 60: 168–177. [Google Scholar]
- Bradshaw W. E., Holzapfel C. M.. 2007. Evolution of animal photoperiodism. Annu. Rev. Ecol. Evol. Syst. 38: 1–25. [Google Scholar]
- Burkett-Cadena N. D. 2011. A wire-frame shelter for collecting resting mosquitoes. J Am. Mosq. Control. Assoc. 27: 152–155. [DOI] [PubMed] [Google Scholar]
- Burkett-Cadena N. D., Graham S. P., Hassan H. K., Guyer C., Eubanks M. D., Katholi C. R., Unnash T. R.. 2008. Blood feeding patterns of potential arbovirus vectors of the genus Culex targeting ectothermic hosts. Am. J. Trop. Med. Hyg. 79: 809–815. [PMC free article] [PubMed] [Google Scholar]
- Burkett-Cadena N. D., Hassan H. K., Eubanks M. D., Cupp E. W., Unnasch T. R.. 2012. Winter severity predicts the timing of host shifts in the mosquito Culex erraticus. Biol. Lett. 8: 567–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett-Cadena N. D., Bingham A. M., Hunt B., Morse G., Unnasch T. R.. 2015. Ecology of Culiseta melanura and other mosquitoes (Diptera: Culicidae) from Walton County, FL, during winter period 2013–2014. J. Med. Entomol. 52: 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craighead J. E., Shelokov A., Peralta P. H.. 1962. The lizard: a possible host for eastern equine encephalitis virus in Panama. Am. J. Hyg. 76: 82–87. [DOI] [PubMed] [Google Scholar]
- Darsie R. F., Ward R. A.. 1981. Identification and geographical distribution of the mosquitoes of North America, north of Mexico. Mosq. Syst. 1S: 1–313. [Google Scholar]
- Edman J. D. 1979. Host-feeding patterns of Florida mosquitoes (Diptera: Culicidae) VI. Culex (Melanoconion). J. Med. Entomol. 15: 521–525. [DOI] [PubMed] [Google Scholar]
- Edman J. D., Taylor D. J.. 1968. Culex nigripalpus: seasonal shift in the bird-mammal feeding ration in a mosquito vector of human encephalitis. Science 161: 67–68. [DOI] [PubMed] [Google Scholar]
- Edman J. D., Webber L. A., Kale H. W. II.. 1972. Host-feeding patterns of Florida mosquitoes II. Culiseta. J. Med. Entomol. 9: 429–434. [DOI] [PubMed] [Google Scholar]
- Florida Department of Health. Mosquito-borne disease surveillance. (http://www.floridahealth.gov/diseases-and-conditions/mosquito-borne-diseases/surveillance.html) (accessed 19 September 2016).
- Graham S. P., Hassan H. K., Chapman T., White G., Guyer C., Unnasch T. R.. 2012. Serosurveillance of eastern equine encephalitis virus in amphibians and reptiles from Alabama, USA. Am. J. Trop. Med. Hyg. 86: 540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C. A., Hedges S. B., Maxson L. R.. 1993. Molecular insights into the relationships and biogeography of West Indian anoline lizards. Biochem. Syst. Ecol. 21: 97–114. [Google Scholar]
- Hayes R. O., Daniels J. B., Maxfield H. K., Wheeler R. E.. 1964. Field and laboratory studies on eastern encephalitis in warm- and cold-blooded vertebrates. Am. J. Trop. Med. Hyg. 13: 595–606. [DOI] [PubMed] [Google Scholar]
- Hosmer D. W., Lemeshow S. Jr, Sturdivant R. X.. 2013. Applied logistic regression, 3rd ed John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- Kilpatrick A. M., Kramer L. D., Jones M. J., Marra P. P., Daszak P.. 2006. West Nile virus epidemics in North America are driven by shifts in mosquito feeding behavior. PLoS Biol. 4: e82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano T., Umetsu K., Tian W., Osawa M.. 2007. Two universal primer sets for species identification among vertebrates. Int. J. Legal Med. 121: 423–427. [DOI] [PubMed] [Google Scholar]
- Lambert A. J., Martin D. A., Lanciotti R. S.. 2003. Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J. Clin. Microbiol. 41: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Tsai L. C., Huang M. T., Jhuang J. A., Yao C. T., Chin S. C., Wang L. C., Linacre A., Hsieh H. M.. 2008. A novel strategy for avian species identification by cytochrome b gene. Electrophoresis 29: 2413–2418. [DOI] [PubMed] [Google Scholar]
- Mahmood F., Crans W. J.. 1998. Effect of temperature on the development of Culiseta melanura (Diptera: Culicidae) and its impact on the amplification of eastern equine encephalomyelitis virus in birds. J. Med. Entomol. 35: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Molaei G., Andreadis T. G.. 2006. Identification of avian-and mammalian-derived bloodmeals in Aedes vexans and Culiseta melanura (Diptera: Culicidae) and its implication for West Nile virus transmission in Connecticut, USA. J. Med. Entomol. 43: 1088–1093. [DOI] [PubMed] [Google Scholar]
- Molaei G., Oliver J., Andreadis T. G., Armstrong P. M., Howard J. J.. 2006. Molecular identification of bloodmeal sources in Culiseta melanura and Culiseta morsitans from an endemic focus of eastern equine encephalitis virus in New York. Am. J. Trop. Med. Hyg. 75: 1140–1147. [PubMed] [Google Scholar]
- Molaei G., Armstrong P. M., Graham A. C., Kramer L. D., Andreadis T. G.. 2015a. Insights into the recent emergence and expansion of eastern equine encephalitis virus in a new focus in the northern New England USA. Parasit. Vectors 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G., Armstrong P. M., Abadam C. F., Akaratovic K. I., Kiser J. P., Andreadis T. G.. 2015b. Vector-host interactions of Culiseta melanura in a focus of eastern equine encephalitis virus activity in southeastern Virginia. PLoS ONE 10: e0136743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G., Thomas M. C., Muller T., Medlock J., Shepard J. J., Armstrong P. M., Andreadis T. G.. 2016. Dynamics of vector-host interactions in avian communities in four eastern equine encephalitis virus foci in the northeastern U.S. PLoS Negl. Trop. Dis. 10: e0004347.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen G. R., Durden L. A.. 2009. Medical and Veterinary Entomology, 2nd Ed Academic Press. [Google Scholar]
- Nagelkerke N.J.D. 1991. A note on a general definition of the coefficient of determination. Biometrika 78: 691–692. [Google Scholar]
- Neill W. T. 1948. Hibernation of amphibians and reptiles in Richmond County, Georgia. Herpetologica 4: 107–114. [Google Scholar]
- Scott T. W., Weaver S. C.. 1989. Eastern equine encephalomyelitis virus: Epidemiology and evolution of mosquito transmission. Adv. Virus Res. 37: 277–328. [DOI] [PubMed] [Google Scholar]
- Tempelis C. H., Reeves W. C., Bellamy R. E., Lofy M. F.. 1965. A three-year study of the feeding habits of Culex tarsalis in Kern County, California. Am. J. Trop. Med. Hyg. 14: 170–177. [DOI] [PubMed] [Google Scholar]
- Vidal N., Kindl S. G., Wong A., Hedges S. B.. 2000. Phylogenetic relationships of xenodontine snakes inferred from 12S and 16S ribosomal RNA sequences. Mol. Phylog. Evol. 14: 389–402. [DOI] [PubMed] [Google Scholar]
- Wellings F. M., Lewis A. L., Pierce L. V.. 1972. Agents encountered during arboviral ecological studies: Tampa Bay area, Florida, 1963 to 1970. Am. J. Trop. Med. Hyg. 21: 201–213. [DOI] [PubMed] [Google Scholar]
- White G. S., Ottendorfer C., Graham S., Unnasch T. R.. 2011. Competency of reptiles and amphibians for eastern equine encephalitis virus. Am. J. Trop. Med. Hyg. 85: 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. S., Kramer L. D., Maffei J. G., Dusek R. J., Backenson P. B., Mores C. N., Bernard K. A., Ebel G. D.. 2008. Molecular epidemiology of eastern equine encephalitis virus, New York. Emer. Infect. Dis. 14: 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]