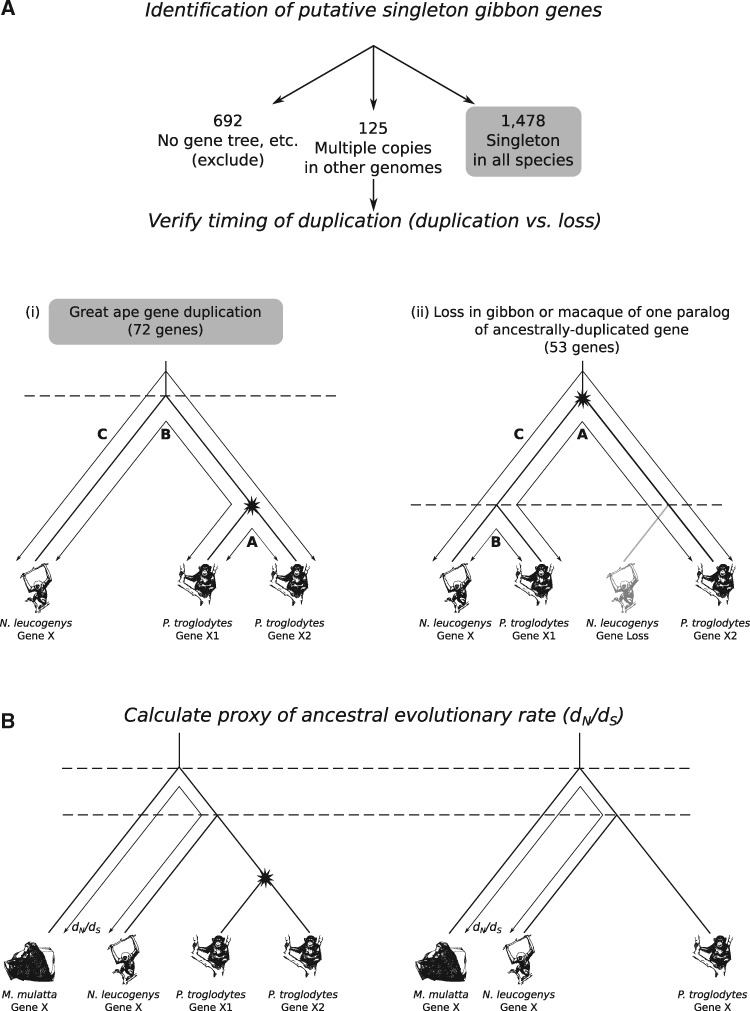
Fig. 1.
Project strategy. We are interested in identifying genes that are singletons in gibbon and macaque, and sorting those into great ape singleton and duplicable genes according to their status in great ape genomes. (a) Compile list of putative singleton gibbon genes (via an all-against-all BLASTp). For each singleton gene, obtain the corresponding Ensembl gene tree. Confirm the singleton status of gibbon and macaque within the trees. Assess their condition (singleton or nonsingleton) in each of the four related primate species human, chimpanzee, gorilla, and orangutan. This restriction to within the primate lineage minimizes the effects of young versus old duplications. The nonsingleton set of gene trees can arise through gene duplication or gene loss events. The two alternative scenarios are illustrated using chimpanzee (panels a—i and ii). If a duplication has occurred in the chimpanzee lineage, the distance, A, between the two sister genes will be less than each of the distances to the orthologous gibbon gene (B and C). However, this relationship will not hold if the nonsingleton arose from a prespeciation duplication event, with a subsequent gene loss in gibbon. This method has been adapted from a similar protocol in He and Zhang (2006). (b) The rate of evolution () in macaque and gibbon is used as a proxy for the ancestral rate of evolution of the primate lineage, independent of any duplication event. Horizontal dashed lines represent speciation events and stars represent gene duplication events.