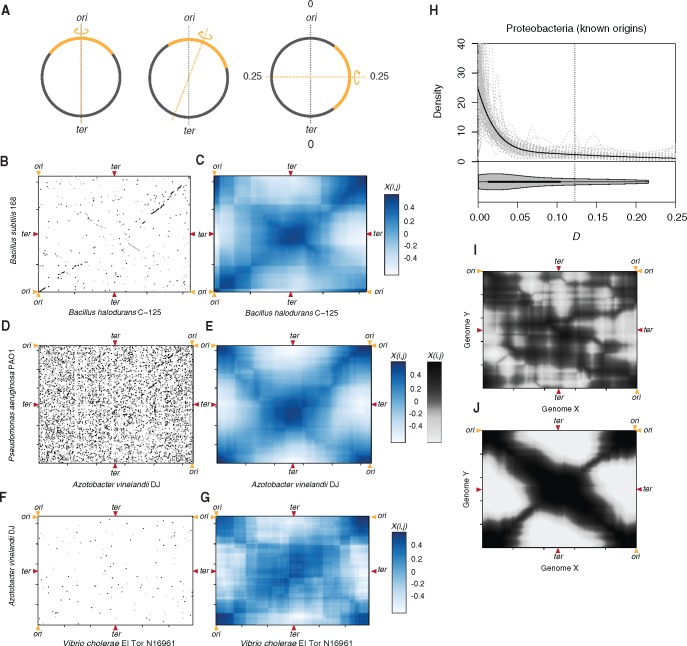
Fig. 1.
Detecting symmetric inversion bias. (A) Inversions in circular prokaryotic genomes can occur with varying degrees of symmetry in relation to the ori-ter axis (grey dotted line). The furthest away the inversion axis (orange dotted line) can be from the ori-ter axis is a quarter of the genome (0.25). (B, D, F) Symmetric inversions cause X-shaped patterns in pairwise MUMmer alignments, which can be hard to discern (D, F) but are revealed as symmetry hot and cold spots when displaying Xi,j (C, E, G). (H) Distribution of D scores for Proteobacteria with experimentally determined ori positions. The black line is the logspline density fit to the whole dataset, the grey lines are logspline density fits to random 40% jack-knifed samples to explore outsize influence of individual genome pairs. (I, J) Xi,j heat maps representing a case of simulated genome divergence by random (I) and symmetric and quasi-symmetric (J) inversions.