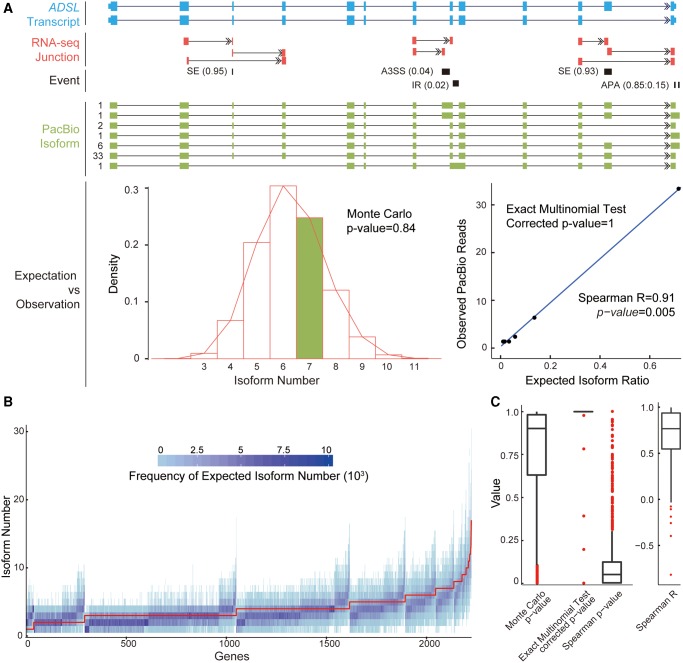
Fig. 3.
Independent combination of alternative RNA processing events in human. (A) Upper panel: as proof-of-concept, the ADSL gene is shown to demonstrate the procedures used to study the combination of alternative RNA processing events. Five alternative RNA processing events were located on the ADSL gene: four AS events and one APA. The inclusion ratio for each AS event, as well as the PA frequency ratio for APA, are shown in brackets. Middle panel: structures of PacBio isoforms with PacBio read coverage highlighted on the left of the isoform structure. Lower left panel: distribution of expected isoform numbers generated by 10,000 iterations of Monte Carlo simulations; green bar indicates the number of isoforms observed in the real PacBio Iso-seq data. Lower right panel: frequency of expected isoforms versus real isoforms in PacBio Iso-seq. (B) Distribution of isoform numbers generated by 10,000 iterations of Monte Carlo simulation shown as a heat-map across each gene (X axis). Red curve: the observed isoform numbers from PacBio Iso-seq. (C) Distribution of Monte Carlo P values for the 2,242 human genes showing whether the observed isoform number was significantly lower than expectation. The distributions of corrected P values from the Exact Multinomial Test, Spearman correlation coefficients, and the Spearman correlation P values are also shown to indicate whether the frequencies of the expected isoforms are consistent with those for the observed isoforms in PacBio sequencing.