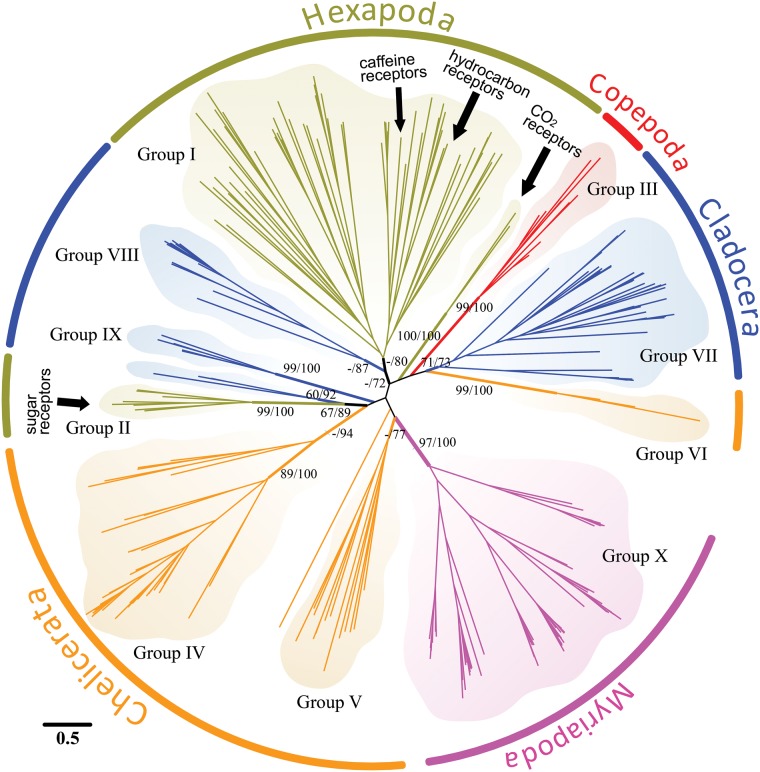
Fig. 2.
Phylogeny of the GR gene families from representatives of some major clades within the Arthropoda. Phylogenetic relationships among GR genes of the fruit fly Drosophila melanogaster (Hexapoda, Groups I and II, olive), the waterflea Daphnia pulex (Cladocera, within Branchiopoda, Groups VII–IX, blue), the copepod Eurytemora affinis (Copepoda, Group III, red), the centipede Strigamia maritima (Myriapoda, Group X, magenta), and the black-legged tick Ixodes scapularis (Chelicerata Groups IV–VI, orange). The phylogeny was constructed using maximum likelihood, based on alignments of 1,346 amino acids of GR genes (see Materials and Methods). The numbers at internal branches show bootstrap support values (%) for the maximum-likelihood reconstruction and posterior probabilities (%) for the Bayesian reconstruction. Support values on the major internal branches are shown for values higher than 60%. Groups I–X each represent lineage-specific expansions (of more than three genes) and are supported by > 0.70 posterior probability in the Bayesian reconstruction. The scale bar represents the number of amino acid substitutions per site. See supplementary figure S1, Supplementary Material online, for a more detailed GR amino acid phylogeny using additional taxa.