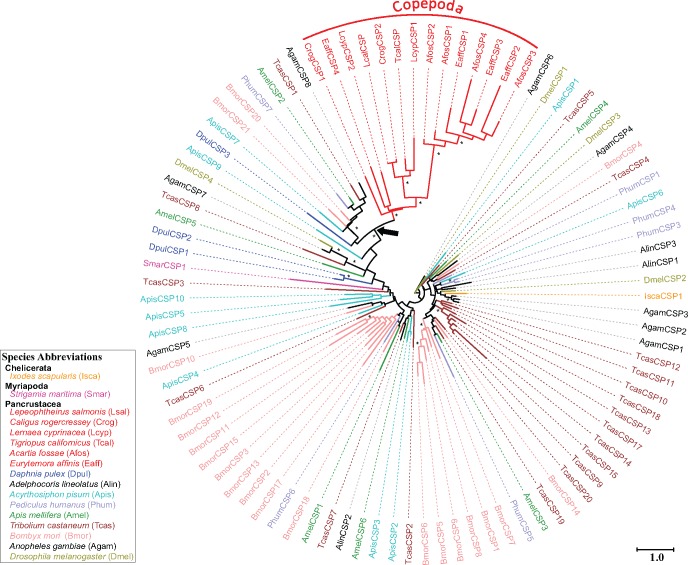
Fig. 6.
Phylogeny of CSPs from 6 copepods and 11 other arthropod species. The phylogeny was constructed using maximum likelihood based on sequence alignments of 402 amino acids. Fourteen CSP sequences from six copepods are included (shown in red). In addition to copepods, 81 CSP sequences are included from 11 representative arthropod species. All amino acid sequences except for the copepod sequences are taken from Vieira and Rozas (2011) and Gu et al. (2012). The asterisks indicate branches with at least one of the phylogenetic reconstruction approaches (maximum-likelihood, neighbor-joining phylogenies, or Bayesian) showing bootstrap values or posterior probabilities greater than 70%. The black arrow on the phylogeny points to the node forming a clade within Pancrustacea, composed of a Daphnia pulex CSP, seven insect CSPs, and copepod CSPs. This clade supports a clear homologous relationship between copepod and insect/branchiopod CSPs. This node is supported by a maximum-likelihood bootstrap value of 62% and a Bayesian posterior probability of 0.78. The following representative species were used in this analysis: the fruit fly Drosophila melanogaster (Diptera, olive), the silkworm moth Bombyx mori (Lepidoptera, pink), the red flour beetle Tribolium castaneum (Coleoptera, brown), the honeybee Apis mellifera (Hymenoptera, dark green), the pea aphid Acyrthosiphon pisum (Hemiptera, cyan), the human body louse Pediculus humanus (Phthiraptera, slate-blue), the waterflea Daphnia pulex (Cladocera, blue), the six copepod species (Copepoda, red), the centipede Strigamia maritima (Myriapoda, magenta), and the black-legged tick Ixodes scapularis (Chelicerata, orange). All other arthropod species are shown in black. The tree is midpoint rooted due to the absence of obvious outgroups. The scale bar represents the number of amino acid substitutions per site.