Figure 2. A pathway for reductive aromatic amino acid metabolism by members of the gut microbiota.
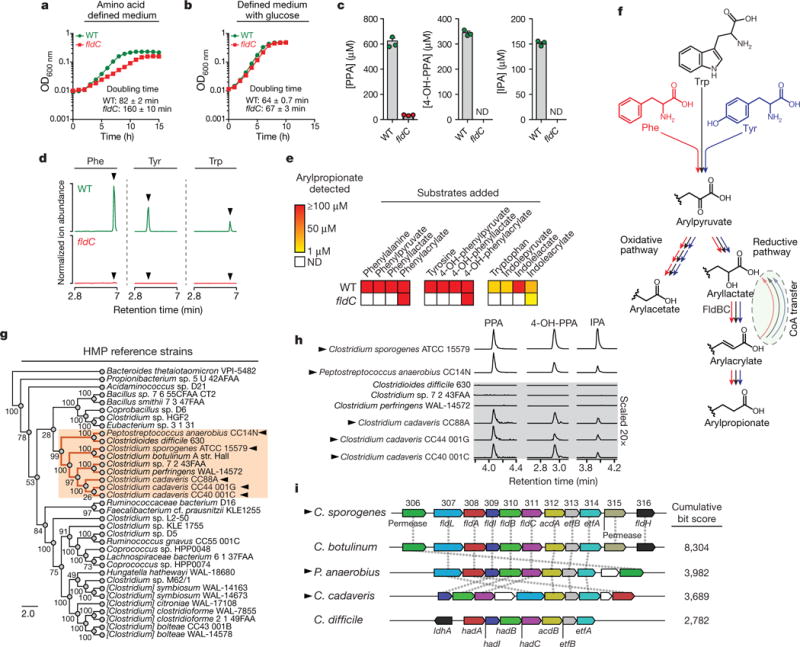
a, b, The fldC mutant exhibits a growth defect during AAA metabolism. OD, optical density; WT, wild type. c, Metabolite analysis of supernatants from cultures in a. d, e, The fldC mutant exhibits a defect in reductive metabolism of AAAs. ND, not detected. f, Summary of the predicted pathway for AAA metabolism showing phenyllactate dehydratase (FldBC). g, Phylogenetic distribution of reference strains. h, Metabolite screening identifies AAA-reducing strains. i, AAA-reducing microbes share similar gene clusters with C. sporogenes. hadA, isocaproyl-CoA:2-hydroxyisocaproate CoA transferase; hadI, 2-hydroxyisocaproyl-CoA dehydratase activator; hadBC, 2-hydroxyisocaproyl-CoA dehydratase; acdB, acyl-CoA dehydrogenase. a, b, d, h, Representative curves from n = 3 biological replicates are shown; a–c, Data are mean ± s.d. Arrowheads in d mark retention times for arylpropionates. Arrowheads in g, h, i indicate strains capable of AAA reduction.
