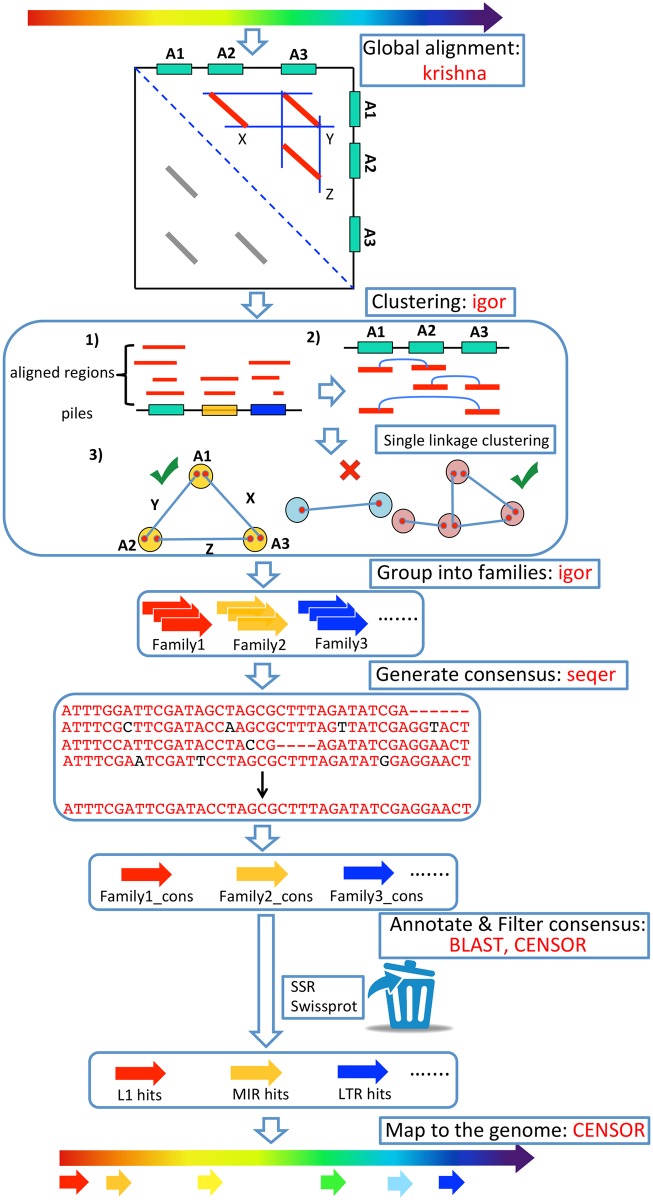
Fig 1. Comprehensive ab initio Repeat Pipeline (CARP).
Figure shows the detailed steps for CARP. Repetitive DNA is identified by all vs all pairwise alignment using krishna. Single linkage clustering is then carried out to produce families of repetitive sequences that are globally aligned to generate a consensus sequence for each family. Consensus sequences are filtered for non-TE protein coding genes and then annotated using Repbase and a custom library of retrovirus and reverse transcriptase sequences. The annotated consensus sequences are then used to annotate the genome. This is required to identify repeats with less than the threshold identity used for alignment that are overlooked during the initial pairwise alignment step.