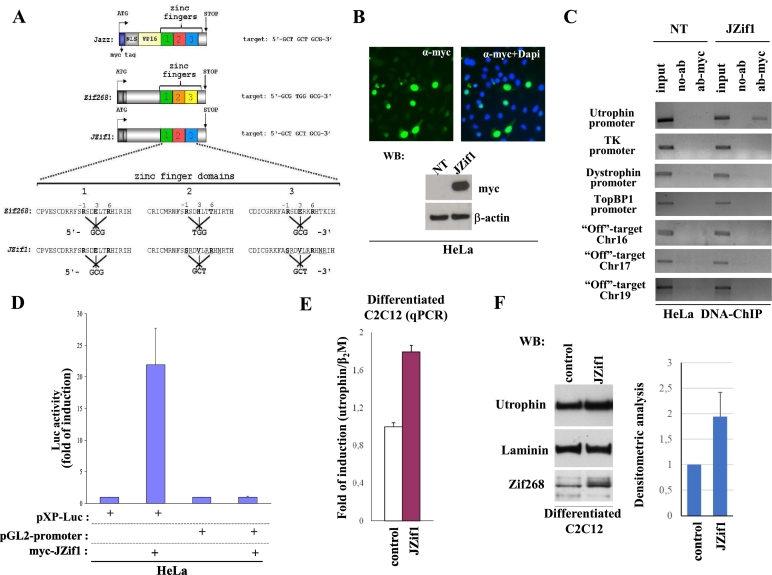
Fig. 1.
The novel artificial transcription factor JZif1.
a. Schematic view of the novel artificial/natural transcription factor JZif1 and its prototype ‘Jazz’. Varied aminoacids in the Zif268 zinc finger region and transcription factors binding sites are indicated. b. Immunofluorescence images showing the nuclear localization of over-expressed myc-JZif1 in HeLa cells (top panel). The fluorescence intensity was normalized against the background signal. Western blot analysis of cell lysates from HeLa cells transiently transfected with myc-JZif1 (bottom panel). c. Chromatin immuno-precipitation (ChIP) was performed in HeLa cells (untransfected or transfected with the myc-JZif1 expression vector) using myc-tag monoclonal antibody or with no antibodies (no-Ab). Immuno-precipitates from each sample were analyzed by PCR performed with primers specific for the human utrophin promoter ‘A’. The PCR of unrelated promoters of thymidine kinase and dystrophin genes were included as controls. The PCR of TopBP1 gene promoter containing Zif268/Egr1 binding sites and the PCR of specific ‘off-target’ regions from chromosome 16, chromosome 17 and chromosome 19 were also included. A sample representing the linear amplification of the total input chromatin (input) was included (lane 1). d. Luciferase activity obtained upon co-transfection among myc-JZif1 and pXP-Luc utrophin promoter ‘A’ construct or the control reporter pGL2-Promoter (Promega) in HeLa cells. The data are presented as the means ± S.D. of three independent experiments that were performed in triplicate. e. Quantitative real time RT-PCR (qPCR) analysis of the utrophin gene mRNA expression in differentiated C2C12 cells transfected with the mAAV-JZif1 or mAAV control vectors. The gene expression ratio of utrophin, normalized as indicated, is shown as the mean ± S.D. from an experiment performed in triplicate. f. Representative Western blot analysis of utrophin protein levels in differentiated C2C12 cells transfected with the mAAV-JZif1 or mAAV control vectors. The antibodies used are indicated. Densitometric analysis represents the mean ± S.D. of four independent experiments (right panel).