Fig. 6.
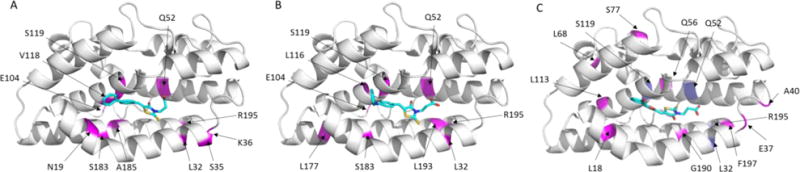
A representation of the pa-HemO based on Protein Data Bank structure (PDB: 1SK7) is shown, indicating residues perturbed upon binding of compounds 18 (A), 21 (B), and 27 (C). (A) In magenta, residues with >3σ chemical shift perturbations listed as the following in increasing order: A84, Q52, L116, L193, L177, E104, S183, R195, L32, and S119. (B) In magenta, residues with >3σ chemical shift perturbations listed as the following in increasing order: K36, Q52, A185, V118, R195, L32, S35, S119, N19, S119 and E104. (C) In magenta, residues with >3σ chemical shift perturbations listed as the following in increasing order: A185, E37, S77, G190, Q56, L113, L68, A40, L18 and F197. In blue, residues L32, Q52, S119 and R195 (shown in purple) disappeared due to the fast exchange rate.
