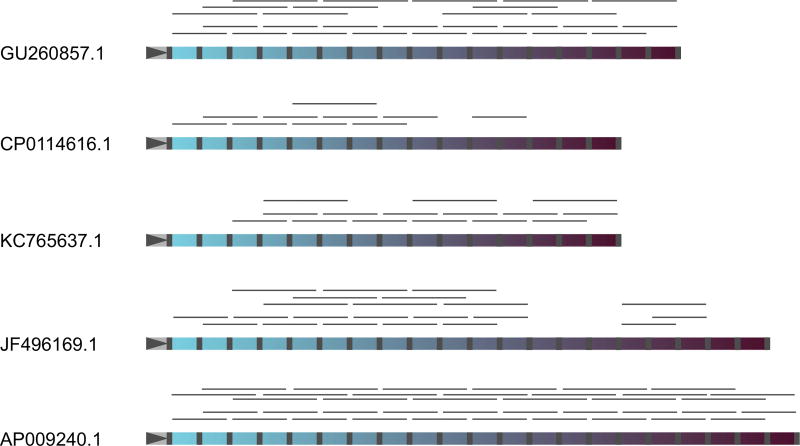
Fig. 3.
Reconstruction of contemporary CRISPR arrays from reads containing two or three spacers from the mammoth sample. Mapping results of neighbouring spacer pairs and triplets on five selected CRISPR arrays from contemporary Escherichia coli are shown. Repeats are grey, and spacers are coloured. The leader regions are marked by grey triangles on the left of each array. Leader-proximal spacers are coloured with lighter shades of blue, while leader-distant spacers are dark-coloured. Detected reads containing neighbouring spacer pairs or triplets are shown by thin grey lines above each array.