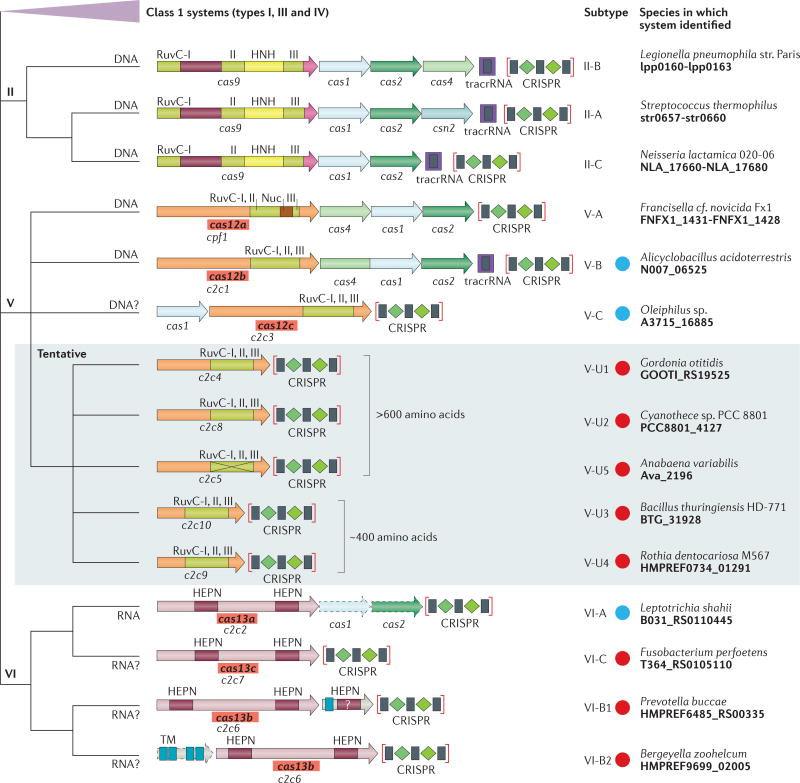
Figure 1. The updated classification scheme for class 2 CRISPR–Cas systems.
The class 1 systems are collapsed; all other systems shown are class 2 systems. New class 2 systems that were discovered using the computational pipeline in this study (see BOX 1) are indicated with blue circles for those that were described previously16 and with red circles for those that are presented here for the first time. For each class 2 system subtype, as well as for the five distinct variants of the provisional V-uncharacterized (V-U) subtype, the locus organization and the domain architecture of the effector and accessory proteins are schematically shown. RuvC-I, RuvC-II and RuvC-III are the three distinct motifs that contribute to the nuclease catalytic centre; numerals in the figure correspond to the respective RuvC motif. The portions of Cas9 proteins that roughly correspond to the recognition lobe and the protospacer-adjacent motif (PAM)-interacting domain are shown by maroon and pink shapes, respectively. The proposed new systematic gene names are shown in bold type in red boxes. Provisional gene names for effector protein candidates are shown below the respective shapes as follows: C2c1–10, class 2 candidate proteins 1–10; for subtype V-A, the previously introduced vernacular cpf1 is indicated. For subtype VI-A, cas1 and cas2 are shown with dashed contours to indicate that only some of these loci include the adaptation module. For the V-U5 variant, the inactivation of the RuvC-like nuclease domain is indicated by a cross. The specific strains of bacteria in which these systems were identified and locus tags for the respective protein-coding genes are also indicated. The abbreviation TM indicates a predicted transmembrane helix. The predicted type of target, namely DNA or RNA, is indicated for each subtype. A question mark next to the target indicates that the activity is only predicted and has not been demonstrated experimentally. The target is not indicated for the type V-U systems because their RNA-guided interference capacity is questionable, which is additionally emphasized by shading. tracrRNA, trans-acting CRISPR RNA.