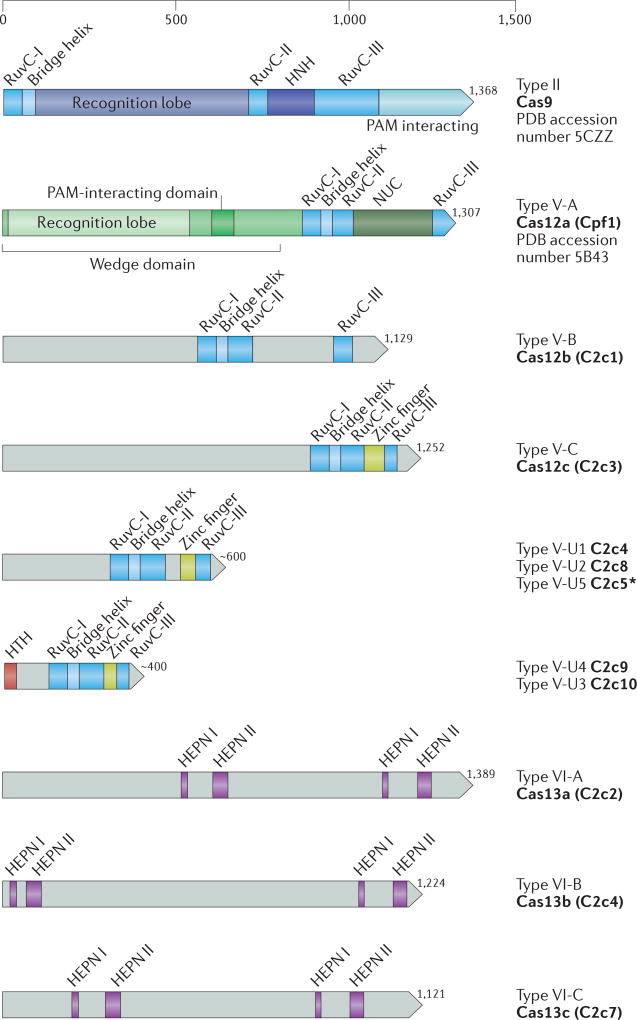
Figure 2. The domain architecture of class 2 CRISPR effector proteins.
For the type II and subtype V-A effectors, the crystal structures (indicated here by their RCSB Protein Data Bank (PDB) accession numbers (5CZZ and 5B43, respectively)) are available and the corresponding domain architectures are shown in detail. For the remainder of the proteins, the grey areas indicate structurally and functionally uncharacterized portions. RuvC-I, RuvC-II and RuvC-III, as well as higher eukaryotes and prokaryotes nucleotide-binding I (HEPN I) and HEPN II, denote the catalytic motifs of the respective nuclease domains of the CRISPR effectors. The bridge helix corresponds to an arginine-rich region that follows the RuvC-I motif. Other domains shown in the figure are denoted as follows: PAM interacting, protospacer-adjacent motif (PAM)-interacting domain; HNH, HNH family endonuclease domain, zinc finger domain with a CXXC.. CXXC motif (dots represent the variable distance between the two pairs of cysteines); HTH, putative DNA-binding helix–turn–helix domain; NUC, nuclease domain. The proteins and domains are shown approximately to scale. For each protein, the corresponding number of amino acids is indicated, and a ruler is shown on top of the figure to guide the eye. For the functionally characterized full-length effectors, the proposed new nomenclature (Cas12 and Cas13) is indicated, whereas for the uncharacterized putative effectors of type V-uncharacterized (V-U), only the provisional names are indicated. When, and if, functional evidence of a bona fide CRISPR response is reported for these effectors, they should be referred to as Cas12 proteins with the corresponding specifying letters. The putative V-U1, V-U2 and V-U5 effectors are larger than the typical TnpB proteins, whereas the V-U3 and V-U4 effectors are in the characteristic size range of TnpB. The asterisk at C2c5 indicates that this putative effector protein contains replacements of the catalytic residues of the RuvC-like nuclease domain and lacks the zinc finger.