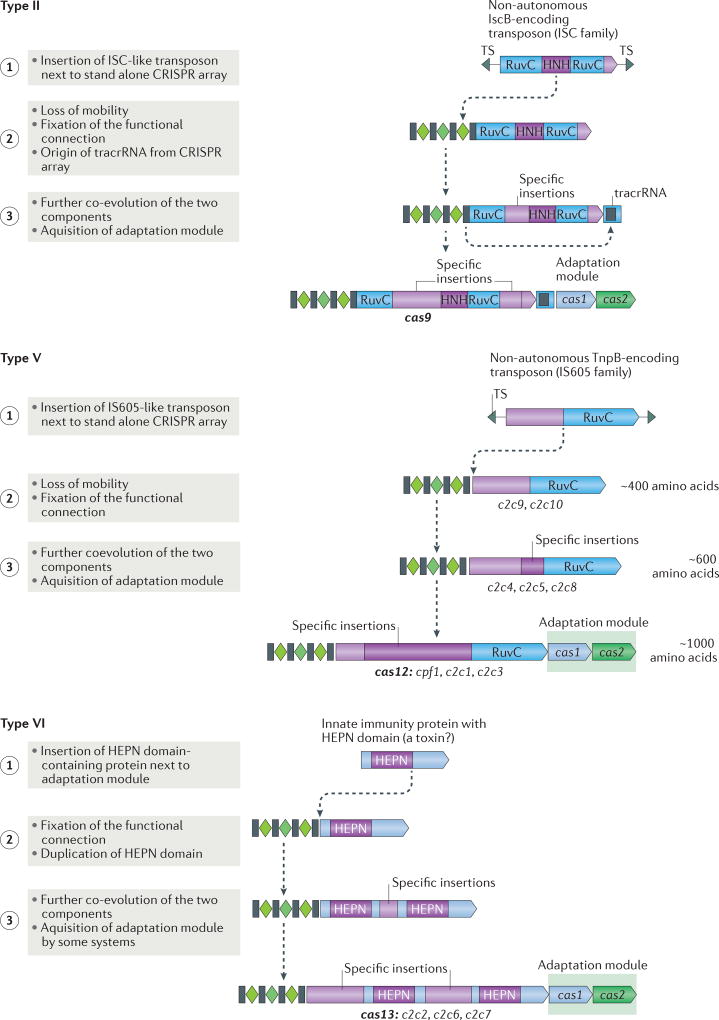
Figure 4. Possible routes of evolution for class 2 CRISPR–Cas systems.
The figure depicts the three-step pathway of the evolutionary ‘maturation’ of type II, type V and type VI CRISPR–Cas systems. The systematic and/or provisional gene names are indicated below the respective ‘mature’ effector protein schematics and the proposed intermediate forms of type V systems. The first step involves the random insertion of a TnpB-encoding or insertion sequences Cas9-like protein B (IscB)-encoding transposon or a higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain RNase-encoding gene next to a CRISPR cassette for type II, type V and type VI systems, respectively. During the second step, the functional connection between this protein and the CRISPR array is established and co-evolution begins, in particular, in the form of the accumulation of specific insertions that facilitate CRISPR RNA (crRNA) binding. For type V systems, the intermediate forms that correspond to the first and second step are identified as different type V-uncharacterized (V-U) variants. Additional components of the system could have originated during the second step, such as trans-acting CRISPR RNA (tracrRNA) in the case of type II systems. During the third step, further insertions lead to increased specificity of crRNA and target binding, and enable interactions with accessory proteins, such as Csn2 for type II-A and a protein with predicted transmembrane (TM) domains for type VI-B. The adaptation module is only inserted into some of the class 2 CRISPR–cas loci during the third step. TS, target site.