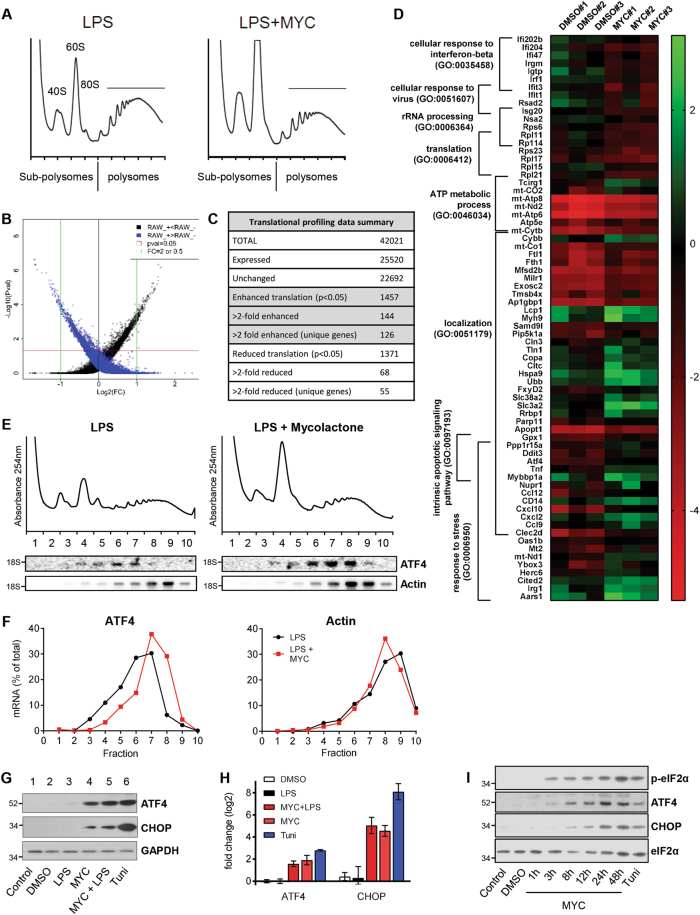
Fig. 1. Translational microarray of cells exposed to mycolactone identifies ATF4 as translationally upregulated.
a−g RAW264.7 cells pre-incubated for 1 h (mycolactone (MYC) or DMSO) then stimulated with 100 ng/ml LPS for 4 h or tunicamycin alone (Tuni). a−e Cell lysates were subject to polysome profiling and translational microarrays, as described in the text. a Mycolactone induces changes in polysome profiles. RNA purified from subpolysomal fractions (1–5) and polysomal fractions (6–10) was pooled and used in microarray analysis as described in Methods. b Scatter plot for probes in the microarray. Black and blue dots represent probes enriched in either polysomes or sub-polysomes respectively. Rank product analysis rules out changes in transcription and achieves a high validation rate for translationally regulated targets. c Summary of microarray data following translational profiling analysis as described in Methods. d Heatmap showing representative data for genes in eight significantly overrepresented gene ontology groups (p < 0.05), identified by PANTHER. e Northern blotting for transcripts in individual gradient fractions from LPS stimulated RAW264.7 cells, the migration of 18S rRNA is indicated; quantified in (f); n = 3 independent experiments. g Cell lysates were analysed by immunoblotting. h Relative fold change (ΔΔCt) for steady-state mRNA levels determined by one-step qRT-PCR on total RNA (Mean ± SEM, n = 3 independent experiments). i HeLa cells were treated as shown and lysates were analysed by immunoblotting. All immunoblots show the approximate migration of molecular weight markers in kDa. See also Figs. S1 and S2, Tables S1 and S2