Abstract
Excessive proliferation and migration of fibroblasts contribute to pulmonary fibrosis in silicosis, and both epithelial cells and endothelial cells participate in the accumulation of fibroblasts via the epithelial–mesenchymal transition (EMT) and the endothelial–mesenchymal transition (EndMT), respectively. A mouse endothelial cell line (MML1) was exposed to silicon dioxide (SiO2, 50 μg/cm2), and immunofluorescence and western blot analyses were performed to evaluate levels of specific endothelial and mesenchymal markers and to elucidate the mechanisms by which SiO2 induces the EndMT. Functional changes were evaluated by analyzing cell migration and proliferation. The mRNA and circular RNA (circRNA) levels were measured using qPCR and fluorescent in situ hybridization (FISH). Lung tissue samples from both Tie2-GFP mice exposed to SiO2 and silicosis patients were applied to confirm the observations from in vitro experiments. Based on the results from the current study, SiO2 increased the expression of mesenchymal markers (type I collagen (COL1A1), type III collagen (COL3A1) and alpha smooth muscle actin (α-SMA/Acta2)) and decreased the expression of endothelial markers (vascular endothelial cadherin (VE-Cad/Cdh 5) and platelet endothelial cell adhesion molecule-1 (PECAM1)), indicating the occurrence of the EndMT in response to SiO2 exposure both in vivo and in vitro. SiO2 concomitantly increased circHECTD1 expression, which, in turn, inhibited HECTD1 protein expression. SiO2-induced increases in cell proliferation, migration, and changes in marker levels were restored by either a small interfering RNA (siRNA) targeting circHECTD1 or overexpression of HECTD1 via the CRISPR/Cas9 system, confirming the involvement of the circHECTD1/HECTD1 pathway in the EndMT. Moreover, tissue samples from SiO2-exposed mice and silicosis patients confirmed the EndMT and change in HECTD1 expression. Our findings reveal a potentially new function for the circHECTD1/HECTD1 pathway and suggest a possible mechanism of fibrosis in patients with pulmonary silicosis.
Introduction
Silicosis is a pulmonary disease characterized by progressive pulmonary fibrosis caused by long-term inhalation of air containing free silica dust. The excessive proliferation and migration of fibroblasts contributes to pulmonary fibrosis in patients with silicosis1,2, and multiple studies have indicated that both epithelial cells and endothelial cells participate in the accumulation of fibroblasts via the epithelial–mesenchymal transition (EMT) and endothelial–mesenchymal transition (EndMT) in different settings3–5.
Although mounting evidence has indicated that both the damage to alveolar epithelial cells and subsequent diffuse inflammatory responses are involved in the pathogenesis of pulmonary fibrosis, the EndMT has received little attention in the context of silicosis. The EndMT occurs in in different organs, such as the kidneys6, liver, and heart7, in patients with fibrotic disorders, as well as in patients with diabetes8, and metastatic tumors7. The EndMT is characterized by the loss of endothelial-specific markers, the acquisition of the mesenchymal or myofibroblast phenotype and the expression of mesenchymal cell products, such as α-smooth muscle actin (α-SMA) and type I collagen (Col I/COL1A1)9.
Noncoding RNAs are involved in the EndMT in different diseases, although the detailed mechanisms remain unclear10–12. Circular RNAs (circRNAs), which are produced by reverse splicing, comprise a new class of noncoding RNAs and have become a hot topic of research in recent years13. circRNAs not only affect mRNA transcriptional levels in the nucleus but also adsorb miRNAs in the cytoplasm or directly interact with specific proteins to affect their transcriptional or post-transcriptional levels13,14. For example, the circRNA ciRS-7 acts as a sponge for miR-7, and ciRS-7 is resistant to miRNA-mediated target destabilization, thus strongly suppressing miR-7 activity15. The circRNA HIPK2 functions as an endogenous microRNA-124 sponge to increase sigma non-opioid intracellular receptor 1 expression16. In addition, circRNAs also affect gene transcription through their associations with phosphorylated Pol II17, and circRNAs can compete with the pre-mRNA splicing machinery18. A recent study from our laboratory based on a circRNA microarray analysis identified 120 circRNAs in the lung that were differentially expressed in silicon dioxide (SiO2)-treated mice compared to normal mice, indicating the fundamental roles of circRNAs in pathological processes induced by SiO2.
In the current study, both circHECTD1 and HECTD1 were involved in the SiO2-induced EndMT by promoting endothelial cell migration and activation. These findings reveal a novel function for circRNAs in SiO2-induced fibrosis and suggest that the circHECTD1/HECTD1 pathway may be involved in multiple steps of the fibrosis process.
Materials and methods
Reagents
SiO2, 80% of which had a particle diameter of less than 5 μm, was purchased from Sigma (S5631), selected via sedimentation according to Stokes’ law, acid hydrolyzed, and baked overnight (200 °C for 16 h)19. The Col I/COL1A1 (BS1530) and type III collagen (Col III/COL3A1, BS1531) antibodies were purchased from BioWord®. The α-SMA/Acta2 antibody (14395-1-AP) was purchased from Proteintech®. The HECTD1 (SC-134976), vascular endothelial cadherin (VE-Cad/Cdh 5, SC-9989), and platelet endothelial cell adhesion molecule-1 (PECAM 1/CD31, SC-1506) antibodies were purchased from Santa Cruz Biotechnology.
Animals
STOCK TEK-GFP 287 Sato/JNju (Tie2-GFP) mice (aged 6–8 weeks, 17–20 g) were obtained from the Laboratory Animal Center of Nanjing Medical University (Nanjing China), and GFP was only expressed in endothelial cells. All animals were males and housed (4 per cage) in a temperature-controlled room (25 °C, 50% relative humidity) with a 12-h light/dark cycle. All animal procedures were performed in strict accordance with the ARRIVE guidelines, and the animal protocols were approved by the Institutional Animal Care and Use Committee of Southeast University.
Cell culture
Mouse microvascular lung (MML1) cells were purchased from Feibo® and maintained in T25 flasks in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS). MML1 cells were stored in liquid nitrogen between passages 3–10 (P3-10). A vial of MML-1 cells was thawed, plated, and passaged until confluence for each experiment, which was performed between passages P5 and P10. Human umbilical vein endothelial cells (HUVECs) were purchased from ScienCell® and maintained in T25 flasks in DMEM supplemented with 10% FBS. HUVECs from passages 3–7 (P3-7) were stored in liquid nitrogen. A vial of P3-7 HUVECs was thawed, plated, and passaged upon reaching confluence to perform each experiment, and each experiment was performed using HUVECs between P10 and P1520.
Establishment of a mouse model of silicosis
Animals were anesthetized with an intraperitoneal injection of pentobarbital sodium, and their tracheae were surgically exposed. A prepared SiO2 suspension (0.2 g/kg in 50 mg/mL saline) was instilled intratracheally in one dose. Lung tissues were collected 28 days after treatment. Control animals were administered the same volume of sterile saline, as previously described21.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cell viability was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays1. Briefly, cells were collected and seeded in 96-well plates at a density of 104 cells/well; the wells on the edges of each 96-well plate were filled with PBS. Then, cells were incubated in a 37 °C incubator with a 5% CO2 atmosphere for 24 h and incubated with 50 μg/cm2 SiO2 for 0, 12, 24, 48, and 72 h. Twenty microliters of MTT dissolved in Hank’s balanced salt solution were added to each well, and the plates were incubated in a 5% CO2 incubator for 1–4 h. Finally, after removing the cell supernatant, cells were treated with 200 μL of dimethyl sulfoxide and agitated on a shaker for 10 min to fully dissolve the formazan crystals. A BioTek microplate reader (SYNERGYH1; BioTek, Highland Park, VT, USA) was used to measure the absorbance of each well at a wavelength of 570 nm. Each experiment was repeated at least three times.
Western blotting
Cells were cultured in 24-well plates in an incubator containing 5% CO2 at 37 °C, and after a variable period of time, cells were washed three times with cold PBS and lysed using a mammalian cell lysis kit (MCL1-1KT, Sigma-Aldrich), according to the manufacturer’s instructions. Equal concentrations of proteins were separated by 8% SDS-PAGE gel electrophoresis under reducing conditions and transferred to PVDF membranes. The PVDF membranes were then blocked with 5% nonfat dry milk in TBST and mildly agitated on a shaker for 1 h at room temperature, followed by incubations with the indicated primary antibodies overnight at 4 °C. Membranes were washed three times with TBS/0.1% Tween 20 for 8 min per wash and incubated with an alkaline phosphatase-conjugated secondary antibody (1:5000 dilution) in 5% nonfat dry milk for 60 min at room temperature. Finally, the PVDF membranes were washed, and the proteins were detected by densitometry using ImageJ software (NIH). Each western blot was repeated at least three times1.
In vitro scratch assay
An in vitro scratch assay was performed to evaluate cell migration in a 2D culture system. Briefly, 1 × 105 MML1 cells were seeded in 24-well tissue culture plates and cultured in growth medium in a 37 °C incubator in a 5% CO2 atmosphere until they reached approximately 70–80% confluence. Then, a sterile 200-µL pipette tip was used to gently scratch the cell monolayer to generate a straight line with a suitable width. Similarly, a second straight line was lightly scratched perpendicular to the first line to create a cross-shaped cellular gap in each well of the 24-well tissue culture plate. Each well was washed twice with fresh growth medium to remove the cell debris, and fresh medium was then added to each well to enable normal cell growth. Simultaneously, each well was incubated with 50 μg/cm2 SiO2, and digital images of the scratches were captured at 0, 12, 36, and 48 h. The ImageJ software was used to quantitatively evaluate the widths of the cell gaps. Each well was examined at least three times1.
Quantitative real-time PCR
The primers are listed in Table S1. TRIzol reagent (Invitrogen) was used to extract the RNA according to the manufacturer’s instructions. Total RNA (mRNA/circRNA) was reverse transcribed using iScript cDNA synthesis kits (Bio-Rad) according to the manufacturer’s instructions. An SsoFast EvaGreen Supermix RT-PCR kit (Bio-Rad) was used to conduct quantitative RT-PCR assays, and the amount of the target RNA was normalized to an endogenous reference (GAPDH) in each experiment. Cycle threshold (Ct) and ΔCT values were analyzed after standardizing the target RNA to the GAPDH RNA. The ΔΔCT quantification method was performed using the Opticon Monitor software (Bio-Rad) to compare the relative expression levels of the treated MML1 cells to the control MML1 cells. The RQ values were determined by assessing the relative fold changes. Quantitative real-time PCR primers were designed using online software and synthesized by GeneChem. Each quantitative real-time PCR analysis was repeated at least three times16.
Immunocytochemistry
Treated cells were washed twice with PBS and fixed with 4% paraformaldehyde in PBS overnight at 4 °C. Then, after two washes, the coverslips were incubated with 0.3% Triton X-100 in PBS at room temperature for 15 min. The permeabilized samples were blocked with 10% NGS in 0.3% Triton X-100 at room temperature for 2 h. Primary antibodies were diluted in PBS containing 10% NGS and 0.3% Triton X-100, and the blocked samples were incubated in primary antibodies overnight at 4 °C. Samples were washed three times and incubated with secondary antibodies at room temperature for 2 h. A mounting solution (Prolong® Gold antifade reagent with DAPI; P36931, Life Technologies) was used to mount the samples after three washes with PBS. Images of the cells were captured using a fluorescence microscope until the coverslips dried. Each experiment was repeated at least three times16.
Fluorescent in situ hybridization (FISH)
Cellular circHECTD1 expression was detected using FISH with a mixture of biotin-labeled DNA oligo probes specific for either endogenous or ectopically expressed circHECTD1. Briefly, cells were freshly fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature, washed twice with PBS, immersed in 70% ethanol overnight at 4 °C, permeabilized with 0.25% Triton X-100 for 15 min, and subjected to two 15-min washes with saline-sodium citrate (SSC) buffer. In situ hybridization was performed overnight at 37 °C using 10 pM biotin-labeled DNA oligo probes in hybridization buffer (HB), and this step was followed by serial washes with SSC buffer. The probe sequence is shown in Table S1. Samples were then incubated in blocking buffer (1% BSA and 3% normal goat serum in PBS) for 1 h at room temperature followed by an anti-biotin HRP antibody (1:200) in blocking buffer overnight at 4 °C. Samples were subsequently subjected to 2-min washes with PBS. Finally, the DNA was stained with DAPI, and images of the cells were captured using a fluorescence microscope (Olympus BX53, Olympus America, Inc., Center Valley, PA, USA).
CRISPR/Cas9 plasmid transfection
The HECTD1 CRISPR activation plasmid (SC-431500-ACT) and control CRISPR activation plasmid (SC-437275) were purchased from Santa Cruz Biotechnology. Approximately 0.5–1 × 105 cells were seeded into 24-well plates (with unparalleled anti-standard medium), and the medium in the 24-well plates was replaced with 200 μL of unparalleled anti-fresh medium per well until the cells reached 40-80% confluence. First, 1.5 μL of the siRNA reagent were added to 10 μL of the transfection medium to form solution A, and the solution was mixed gently at room temperature for 5 min. Second, 0.3 μg of the plasmid was added to 10 μL of the transfection medium to form solution B, and the solution was mixed gently at room temperature for 5 min. Third, solution B was added dropwise to solution A to form solution C, which was immediately vortexed at room temperature and incubated for ≥ 20 min. Finally, solution C was added dropwise to the wells of the 24-well plate, followed by mixing. 12 h after transfection, 1 mL of medium was added to the medium in each well of the 24-well plate, and the medium was discarded and replaced with 1 mL of fresh medium, if the cells were in good condition. The samples were incubated for 24–72 h in a 37 °C incubator containing 5% CO2 until use in the western blotting analysis16.
RNA interference targeting circHECTD1 using siRNAs
RNA interference targeting circHECTD1 was performed in MML1 cells using a previously described method16, with some modifications. The RNA interference protocol was applied to each well of a 24-well plate. Briefly, 3 μL of the siRNA reagent in serum-free DMEM were added to 10 μL of medium to form solution A, and the solution was mixed gently at room temperature for 5 min. Then, 3 μL of the plasmid in serum-free DMEM were added to 10 μL of medium to form solution B, and the solution was mixed gently at room temperature for 5 min. Then, solution B was added dropwise to solution A to form solution C and incubated at room temperature for ≥20 min. Next, 80 μL of serum-free DMEM were added to solution C to form a 100-µL system solution. Cells were washed with PBS and digested with trypsin, and the appropriate amount of normal medium was added to stop the digestion. Next, 100 μL of a 2 × 106 cells/mL cell suspension was added to the above 100-µL system solution to form a 200-µL system solution. The configured 200-µL system solution was added to the wells of a 24-well plate and incubated in a 37 °C incubator containing 5% CO2 for 24–72 h until use in the western blotting analysis.
Nested matrix model and cell migration assay
A nested collagen matrix model was performed as previously described22, with certain modifications. The nested collagen matrix was incubated in an attached state with in DMEM containing 10% FBS for 72 h. The matrix was then removed from the culture well, and 60 µL of a fresh acellular collagen matrix solution (NeoMatrix solution) were added to the center of a new well in a 12-well plate. The newly transferred matrix was covered with 140 µL of the NeoMatrix solution and allowed to polymerize for 1 h at 37 °C in a 5% CO2 atmosphere. Then, 2 mL of DMEM containing 10% FBS were added to the well. Cell migration out of the nested matrix and into the acellular NeoMatrix was quantified by fluorescence microscopy after 24 h of nesting. Digital images (constant dimensions of 1000 × 800 µm) were captured using an EVOS® FL Cell Imaging microscope (Life Technologies, Grand Island, NY, USA) from 3 to 5 randomly selected microscopic fields at the interface of the nested matrix and acellular NeoMatrix. Migration of MML1 cells was quantified by counting the number of cells that had clearly migrated out of the nested matrix into the cell-free matrix. The maximum migration distance was quantified by identifying the cells that had migrated the greatest distance into the cell-free matrix. The number of cells that had migrated out of the nested matrix per field was also quantified from the digital micrographs.
Sirius scarlet stains
For the histological analysis of pulmonary fibrosis, the lung was extracted, fixed with 4% formalin and dehydrated in a 30% sucrose solution. Sections of the lung were examined using a Sirius red staining kit (ab150681, Abcam), according to the manufacturer’ s instructions21.
Statistics
Data are presented as the means ± standard errors of the means. Unpaired numerical data were compared using an unpaired t-test (two groups) or analysis of variance (more than two groups). A P-value <0.05 was regarded as statistically significant.
Results
SiO2 induces the EndMT
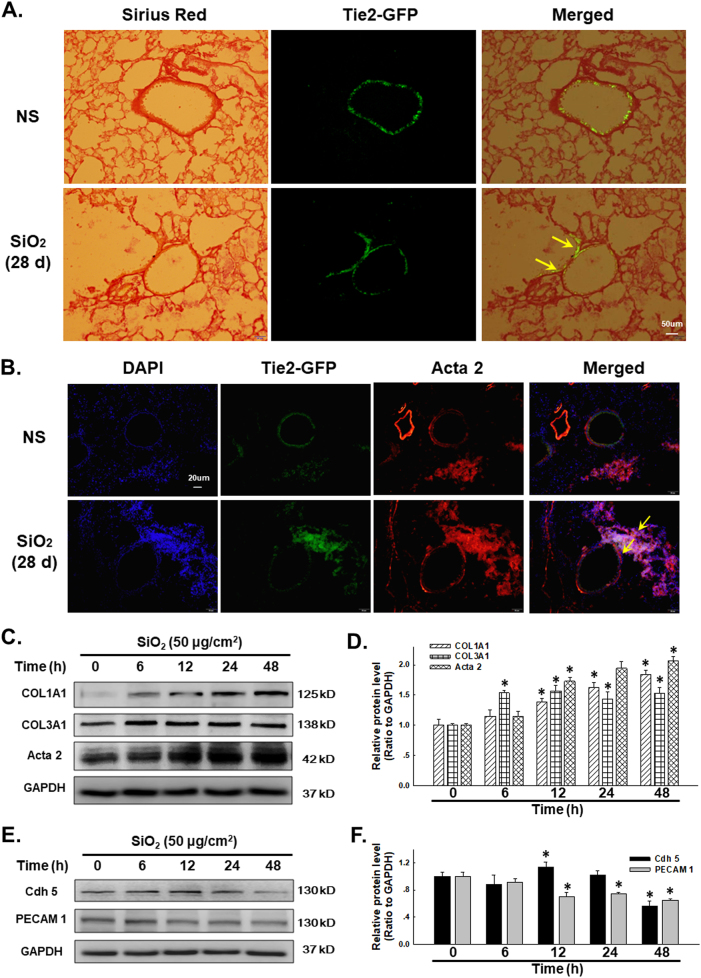
According to a previous study from our laboratory, SiO2 induces the EndMT in HUVECs1. However, researchers have not clearly determined whether SiO2 also induces the EndMT in vivo. In the current study, Tie2-GFP mice were analyzed to determine whether SiO2 induced an EndMT response in the lungs of these mice using the Sirius red staining assay, which enabled differential measurements of the amount of collagen deposited in endothelial cells. The representative images of Sirius red staining in lung sections shown in Fig. 1a, S1a–b show the amount of Sirius red-stained collagen that co-localized with GFP after the SiO2 treatment. Meanwhile, as shown in Fig. 1b, GFP co-localized with α-SMA/Acta2, indicating that SiO2 induced the EndMT. MML1 was used to confirm the in vivo observations. As shown in Fig. 1c, d, SiO2 induced a rapid and sustained increase in Col I/COL1A1, Col III/COL3A1 and α-SMA/Acta2 levels, which are specific mesenchymal markers, indicating the transformation from the endothelial phenotype; however, levels of VE-Cad/Cdh-5 and PECAM 1/CD31, which are specific endothelial markers, showed a delayed and sustained decrease in response to the SiO2 treatment (Fig. 2e, f). The changes in both the endothelial and mesenchymal markers were confirmed by immunostaining (Fig. 2a).
Fig. 1. SiO2 exposure induces the EndMT in vivo and in vitro.
a Representative images of Sirius red staining in lung sections show the amount of Sirius red-stained collagen associated that co-localized with Tie2-GFP in mice treated with SiO2. b Representative images of immunohistochemical staining show the co-localization of TEK-GFP with α-SMA/Acta2 in TEK-GFP 287 Sato/JNju mice treated with SiO2, indicating that SiO2 induced the EndMT. c Representative western blot showing the effects of SiO2 (50 μg/cm2) on levels of the mesenchymal markers Col I/COL1A1, Col III/COL3A1 and α-SMA/Acta2. d Densitometric analyses of five separate experiments show that SiO2 increases Col I/COL1A1, Col III/COL3A1 and α-SMA/Acta2 expression in a time-dependent manner. *P < 0.05 vs. the expression of the corresponding protein at 0 h. e Representative western blot showing the effects of SiO2 (50 μg/cm2) on levels of the endothelial markers VE-Cad/Cdh-5 and PECAM 1/CD31. f Densitometric analyses of five separate experiments show that SiO2 decreases VE-Cad/Cdh-5 and PECAM 1/CD31 expression in a time-dependent manner. *P < 0.05 vs. corresponding protein at 0 h
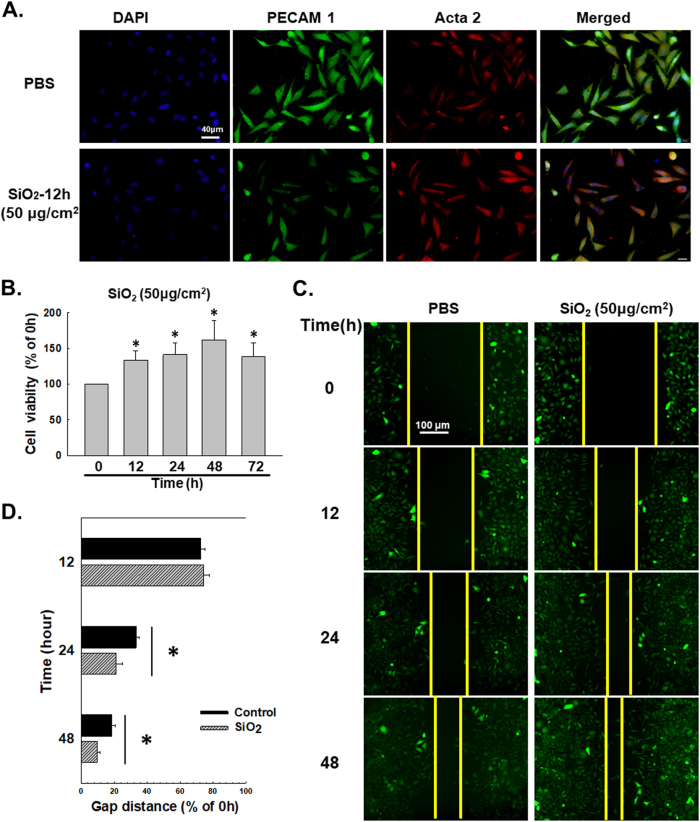
Fig. 2. SiO2 induces the proliferation and migration of MML1 cells.
a Representative images of immunocytochemical staining show that SiO2 (50 μg/cm2) decreases PECAM 1/CD31 expression and increases α-SMA/Acta2 expression in MML1 cells. b MTT assay showing the SiO2-induced increase in the viability of MML1 cells. *P < 0.05 vs. the 0-h group, n = 5. c Representative images showing the effects of SiO2 on GFP-labeled MML1 cell migration in scratch assays. Scale bar = 80 µm. d Quantification of the scratch gap distances in six separate experiments. *P < 0.05 vs. the control group at the corresponding time point
SiO2 induces MML1 cell proliferation and migration
Based on accumulating evidence, changes in cell proliferation and migration comprise the onset of the EndMT and pulmonary fibrosis23–26. Cell migration and viability were evaluated following exposure to SiO2 to further determine the functional effects of endothelial marker loss and mesenchymal marker acquisition by these cells. As shown in Fig. 2b, SiO2 exposure increased cell viability. Moreover, cell migration began to increase after 24 h of SiO2 exposure (Fig. 2c, d), indicating that functional changes were associated with changes in the endothelial and mesenchymal markers.
SiO2 induces circHECTD1 expression in MML1 cells
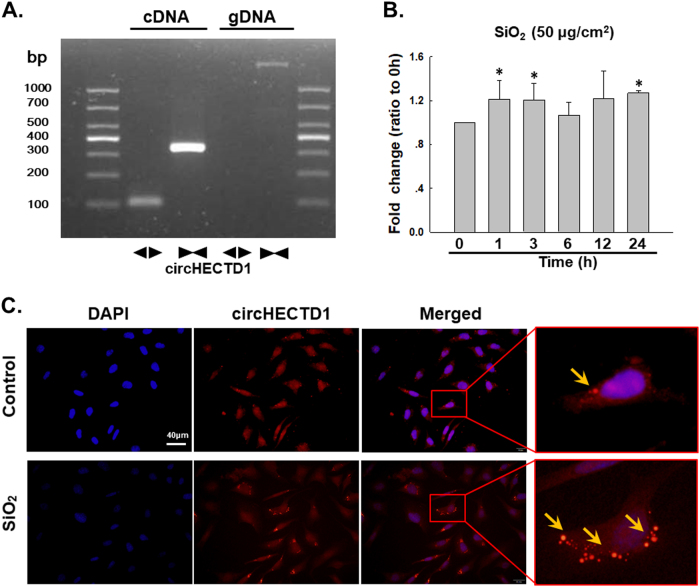
Based on recent reports, circRNAs are involved in different diseases27–31, but researchers have not clearly determined whether circRNAs are involved in the development of silicosis. A recent circRNA microarray analysis-based study revealed 120 differentially expressed circRNAs in the lungs of silicosis mice (Figure S2), among which circHECTD1 is of particular interest because its host gene, HECTD1, a candidate E3 ubiquitin ligase, may be involved in SiO2-induced fibrosis by inducing ubiquitination32. Based on previous data from our laboratory, ZC3H12A/MCPIP1 may mediate macrophage activation and fibroblast proliferation/migration by inducing ubiquitination19,33,34, but the detailed mechanism is unknown. Thus, the effect of SiO2 on circHECTD1 expression was measured in MML1 cells. First, the amplification of circHECTD1 from the cDNA, but not the genomic DNA (gDNA), was confirmed using divergent primers (Fig. 3a). The sequence of circHECTD1 is shown in Figure S3 and Table S2. SiO2 induced a rapid increase in circHECTD1 expression in MML1 cells (Fig. 3b), which was confirmed using FISH (Fig. 3c). Moreover, SiO2 also induced circHECTD1 expression in human endothelial cell line-HUVECs (Figure S4).
Fig. 3. SiO2 induces circHECTD1 expression in MML1 cells.
a Divergent primers amplified circRNAs from cDNAs, but not from gDNA. b As shown in the qRT-PCR analysis, circHECTD1 expression increased in the MML1 cells exposed to SiO2 (n = 5). *P < 0.05 vs. circHECTD1 expression at 0 h. c FISH analysis showing that circHECTD1 expression increases in MML1 cells exposed to SiO2. circHECTD1 was labeled with fluorescein isothiocyanate
circHECTD1 mediates the SiO2-induced EndMT
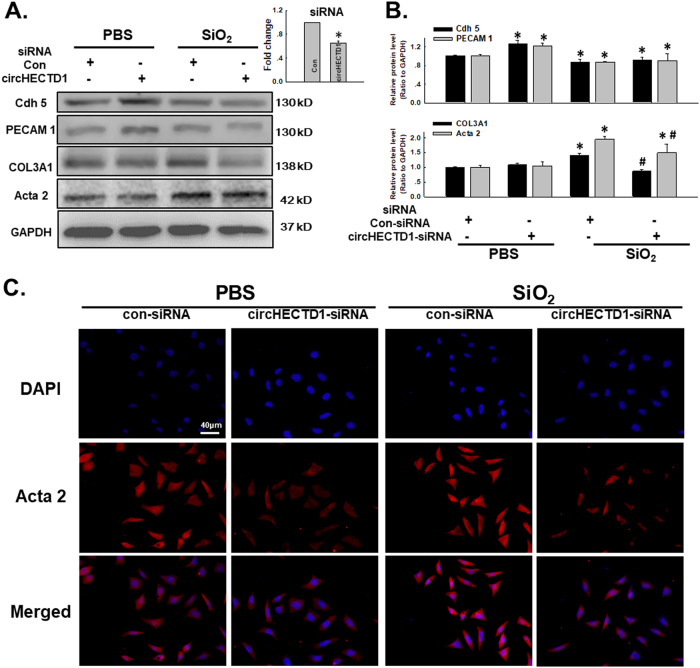
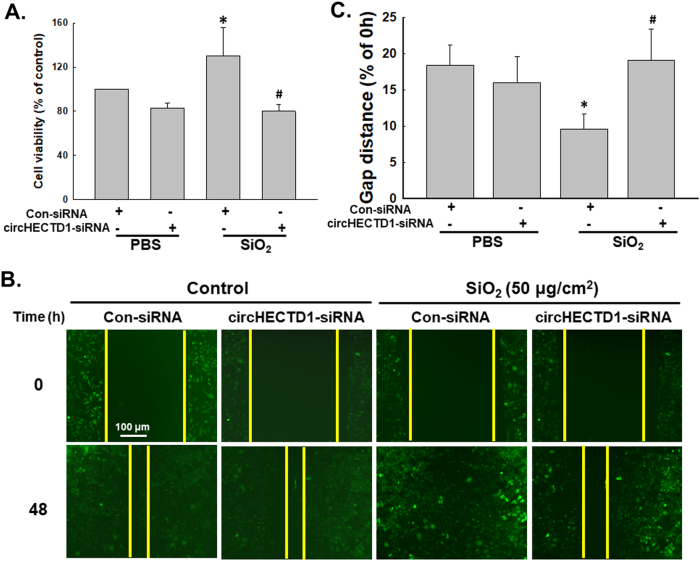
Because SiO2 affected circHECTD1 expression in endothelial cells, we aimed to clarify whether circHECTD1 was involved in the SiO2-induced EndMT. As shown in Fig. 4a, b, specific knockdown of circHECTD1 using siRNAs significantly inhibited the SiO2-induced decreases in VE-Cad/Cdh-5 and PECAM 1/CD31 expression and the increases in Col III/COL3A1 and α-SMA/Acta2 expression, which were confirmed by immunostaining (Fig. 4c). Meanwhile, the SiO2-induced increases in endothelial cell viability and migration were also significantly reversed after circHECTD1 knockdown (Fig. 5a, c).
Fig. 4. circHECTD1 mediates the SiO2-induced EndMT.
a Representative western blot showing the effect of specific knockdown of circHECTD1 with siRNAs on SiO2-induced changes in endothelial and mesenchymal marker expression. The small pane shows the efficacy of circHECTD1-siRNA in reducing circHECTD1 expression. *P < 0.05 vs. the control group, n = 5. b Densitometric analyses of five separate experiments suggest that the SiO2-induced changes in the levels of endothelial and mesenchymal markers were attenuated by the circHECTD1-siRNA. *P < 0.05 vs. the level of the corresponding protein in the control group; #P < 0.05 vs. the level of the corresponding protein in the SiO2 group. c Representative images of immunocytochemical staining show that SiO2-induced α-SMA/Acta2 expression was attenuated by circHECTD1-siRNA
Fig. 5. circHECTD1 is involved in SiO2-induced MML1 cell proliferation and migration.
a MTT assay showing that circHECTD1-siRNA attenuates the SiO2-induced increase in MML1 cell viability. *P < 0.05 vs. the control group; #P < 0.05 vs. the SiO2 group, n = 5. b Representative images show that the effect of SiO2 on GFP-labeled MML1 cell migration in scratch assays was attenuated by circHECTD1-siRNA. Scale bar = 80 µm. c Quantification of the scratch gap distances in six separate experiments. *P < 0.05 vs. the control group; #P < 0.05 vs. the SiO2 group
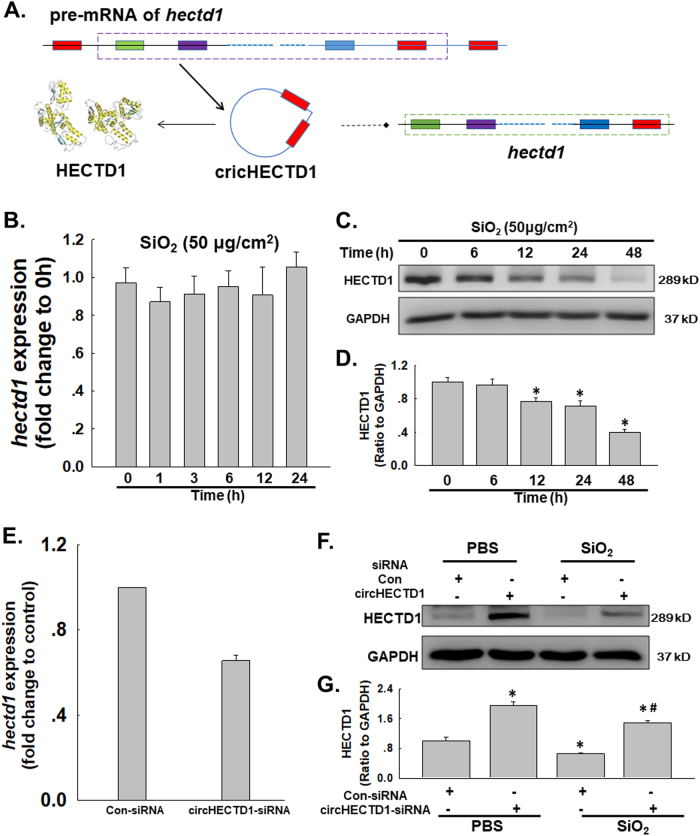
circHECTD1 is involved in SiO2-induced HECTD1 downregulation
After determining the role of circHECTD1 in the SiO2-induced EndMT, we investigated the involvement of its host gene, HECTD1 (Fig. 6a). As shown in Fig. 6b–d, the expression of the hectd mRNA in MML1 cells was not altered after SiO2 exposure, whereas the level of the HECTD1 protein decreased in a time-dependent manner. Moreover, although specific knockdown of circHECTD1 induced a decreasing trend in the expression of the hectd1 mRNA, the circHECTD1-siRNA not only increased the level of the HECTD protein in the normal MML1 cells but also restored the HECTD1 level in MML1 cells exposed to SiO2 (Fig. 6e–g), indicating a role for HECTD1 in the SiO2-induced EndMT.
Fig. 6. circHECTD1 is involved in SiO2-induced HECTD1 downregulation.
a In the SiO2-induced EndMT, circHECTD1 may regulate the expression of its host gene, hectd1. b As shown in the qRT-PCR analysis, levels of the hectd1 mRNA did not change in MML1 cells exposed to SiO2 (n = 5). c Representative western blot showing the effect of SiO2 on the level of the HECTD1 protein. d Densitometric analyses of five separate experiments suggest that SiO2 increases the level of the HECTD1 protein in MML1 cells. *P < 0.05 vs. protein expression at 0 h. e As shown in the qRT-PCR analysis, the expression of the hectd1 mRNA decreased in MML1 cells treated with circHECTD1-siRNA. *P < 0.05 vs. the control group, n = 5. f Representative western blot showing the effect of specific knockdown of circHECTD1 with siRNAs on SiO2-induced HECTD1 expression. g Densitometric analyses of five separate experiments suggest that SiO2-induced changes in HECTD1 expression are attenuated by circHECTD1-siRNA. *P < 0.05 vs. the levels of the corresponding protein in the control group; #P < 0.05 vs. the levels of the corresponding protein in the SiO2 group
HECTD1 is involved the SiO2-induced EndMT in MML1 cells
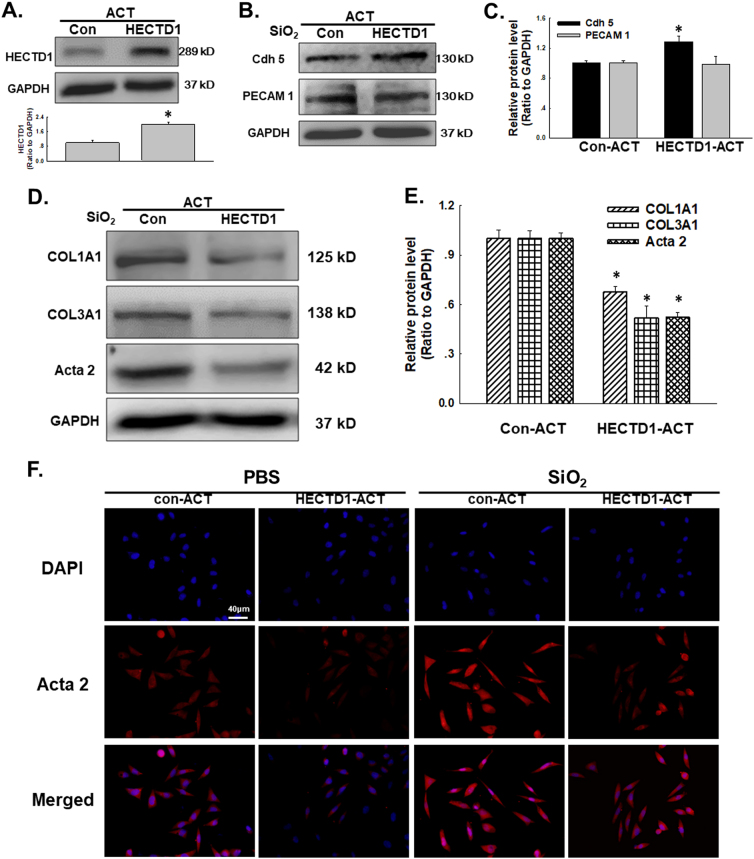
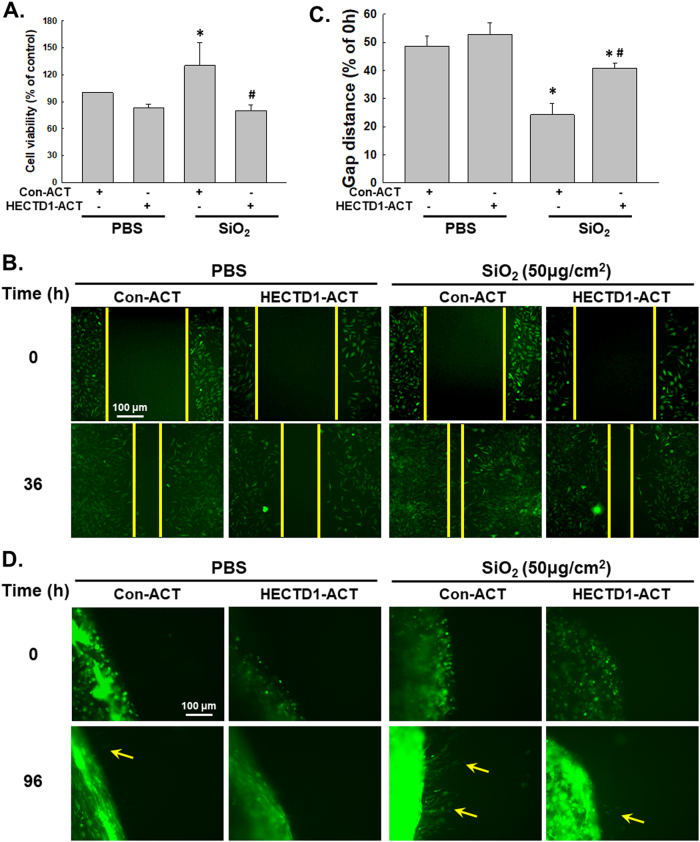
Functional experiments were performed to investigate the effects of HECTD1 on cell proliferation and migration and to obtain a better understanding of the role of HECTD1 in the SiO2-induced EndMT. As shown in Fig. 7a, the HECTD1-CRISPR activation plasmid (ACT) significantly increased the HECTD1 levels in MML1 cells. Meanwhile, upregulation of HECTD1 in the cells restored the SiO2-induced decrease in VE-Cad/Cdh-5 and PECAM 1/CD31 (Fig. 7b, c) levels and the SiO2-induced increase in Col I/COL1A1, Col III/COL3A1 and α-SMA/Acta2 levels (Fig. 7d, e). This effect was confirmed using immunostaining (Fig. 7f). Moreover, the increases in endothelial cell viability and migration induced by SiO2 were also significantly reversed by the HECTD-ACT treatment (Fig. 8a, c). Additionally, the nested matrix cell migration assay was performed to confirm the role of HECTD1 in cell migration. As shown in Fig. 8d, the SiO2-induced increase in cell migration was attenuated by the HECTD-ACT treatment, suggesting a role for HECTD1 in the SiO2-induced EndMT.
Fig. 7. HECTD1 is involved in the SiO2-induced EndMT in MML1 cells.
a Representative western blot and densitometric analyses showing the efficacy of HECTD1 CRISPR ACT at increasing the level of the HECTD1 protein. *P < 0.05 vs. the control group, n = 5. b Representative western blot showing the effect of specific upregulation of HECTD1 expression with ACT on SiO2-induced endothelial marker expression. c Densitometric analyses of five separate experiments suggest that the SiO2-induced changes in VE-Cad/Cdh-5 levels, but not PECAM 1/CD31 levels, are attenuated by HECTD1 ACT. *P < 0.05 vs. the level of the corresponding protein in the control group; #P < 0.05 vs. the level of the corresponding protein in the SiO2 group. d Representative western blot showing the effect of specific upregulation of HECTD1 with ACT on SiO2-induced mesenchymal marker expression. e Densitometric analyses of five separate experiments suggest that the SiO2-induced changes in mesenchymal markers are attenuated by HECTD1 ACT. *P < 0.05 vs. the level of the corresponding protein in the control group; #P < 0.05 vs. the level of the corresponding protein in the SiO2 group. f Representative images of immunocytochemical staining show that SiO2-induced α-SMA/Acta2 expression was attenuated by HECTD1 ACT
Fig. 8. HECTD1 is involved in SiO2-induced MML1 cell proliferation and migration.
a MTT assay showing that HECTD1 ACT attenuated the SiO2-induced increase in MML1 cell viability. *P < 0.05 vs. the control group; #P < 0.05 vs. the SiO2 group, n = 5. b Representative images show that the effect of SiO2 on GFP-labeled MML1 cell migration in scratch assays was attenuated by HECTD1 ACT. Scale bar = 80 µm. c Quantification of the scratch gap distances in six separate experiments. *P < 0.05 vs. the control group; #P < 0.05 vs. the SiO2 group. d Representative images show that the effect of SiO2 on GFP-labeled MML1 cell migration in the nested matrix cell migration assay was attenuated by HECTD1 ACT. Scale bar = 80 µm
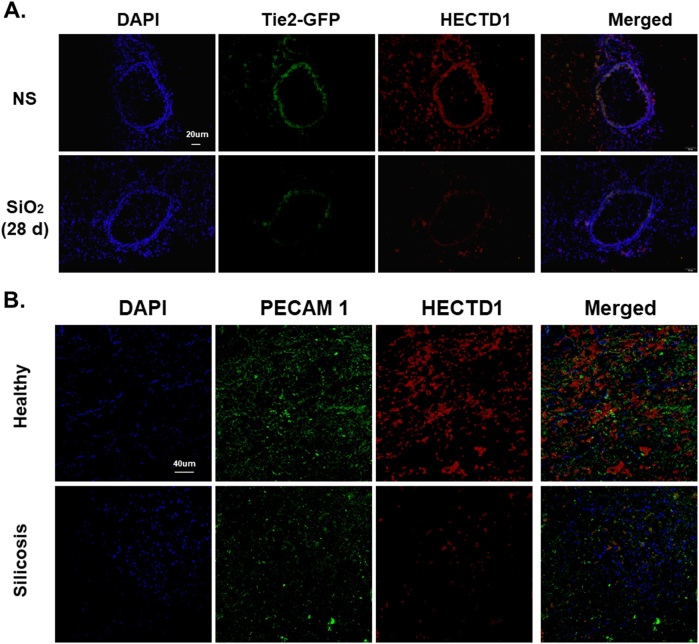
HECTD1 is involved the EndMT in vivo
HECTD1 levels were evaluated in lung tissue samples from both mice and patients to validate our in vitro findings. As shown in Fig. 9a HECTD1 did not co-localize with GFP in normal mice after the SiO2 treatment. Lung tissues from the healthy donors and patients with silicosis showed decreases in PECAM 1/CD31 and HECTD1 staining, which supported our in vitro findings (Fig. 9b).
Fig. 9. HECTD1 is involved in the EndMT in vivo.
a Representative images of immunohistochemical staining show that Tie2-GFP co-localization with HECTD1 decreased in TEK-GFP 287 Sato/JNju mice treated with SiO2. b Representative images of immunohistochemical staining show the expression patterns of PECAM 1/CD31 and HECTD1 in lung sections from healthy donors and patients with silicosis
Discussion
Pulmonary fibrosis is a key and late response of silicosis, and no effective therapies or drugs are currently available to prevent or minimize this progression2,35. In addition to fibroblasts, both epithelial cells and endothelial cells contribute to fibrosis via the EMT and EndMT, respectively, and the detailed mechanisms remain unclear3–5. Compared to the EMT, which has been widely studied, the EndMT has received little attention in fibrotic diseases. The current study mainly focused on the roles of circRNAs and their downstream effects on the SiO2-induced EndMT, revealing a new strategy for treating silicosis.
SiO2 destroys the barrier between the air and blood, which exposes endothelial cells to silica36. This disruption results in not only activation of endothelial cells but also the EndMT, which exacerbates endothelial cell dysfunction1,4. The EndMT is characterized by the loss of an endothelial phenotype, the acquisition of a mesenchymal phenotype, and functional changes in the cell, including proliferation and migration1,9. Similar to the EMT, the EndMT is induced by transforming growth factor (TGF-β)9. For example, endothelial cell-derived fibroblasts participate in bleomycin-induced lung fibrosis, which is attenuated following the withdrawal of TGF-β both in vitro and in vivo4. Interestingly, hypoxia, a common feature of many diseases, plays an important role in various fibrotic diseases, including SSc, kidney, cardiac, and pulmonary fibrosis37. For example, hypoxia-induced HIF-1α expression promotes the EndMT during the development of radiation-induced pulmonary fibrosis38, indicating a role for the lung as a source triggering the EndMT in subjects with fibrotic diseases. Despite the importance of the EndMT during experimentally induced pulmonary fibrosis4, no reports have described the EndMT in patients with idiopathic pulmonary fibrosis or other forms of secondary pulmonary fibrosis37; however, the current study has provided another example of the EndMT in patients with pulmonary fibrosis.
Although the mechanism underlying the EndMT in pulmonary fibrosis deserves further investigation, several studies of cardiac fibrosis associated with the EndMT have indicated a role for noncoding RNAs in this pathological process. For example, the EndMT observed in the hearts of wild type diabetic mice is prevented in miR-200b transgenic diabetic mice, suggesting that miR-200b exerts a protective effect by inhibiting TGF-β1 and p300 expression11. Interestingly, miR-200a negatively regulates levels of the GRB2 protein by directly binding to the grb2 3′UTR, which, in turn, inhibits the EndMT in a cardiac interstitial fibrosis model39. Unlike miRNAs, which have been widely studied, circRNAs comprise a newly identified and highly abundant RNA species that has received increasing attention in recent decades. The circular structure of circRNAs is associated with high biochemical stability and conservation40, making circRNAs ideal candidate biomarkers for the diagnosis and targeted treatment of diseases41. Recent studies have suggested the potential use of circRNAs in the diagnosis and treatment of nervous system disorders and cancer16,42–44. For example, circHIPK2 promotes astrocyte and fibroblast activation through a ceRNA-mediated mechanism, in which circHIPK2 functions as an endogenous miRNA sponge, resulting in increased SIGMAR1 expression16,45. Based on our previous circRNA microarray data and EndMT studies32, we selected circHECTD1, which promoted the EndMT in vitro after SiO2 exposure, in the current study. Unlike the classical ceRNA mechanism, circHECTD1 may regulate the protein level of its host gene, hectd1, through competition with its pre-mRNA, although further experiments are needed to confirm this hypothesis. The involvement of circHECTD1 in the SiO2-induced EndMT highlights circRNAs as new potential targets for silicosis treatment.
Although the detailed mechanism underlying the effects of circHECTD1 on the EndMT requires further investigation, circHECTD1 promotes functional changes in cells through HECTD1. HECTD1 is an E3 ubiquitin ligase that contains an N-terminal ankyrin repeat, a MIB domain, and a C-terminal HECT domain, which determines target protein specificity46; HECTD1 plays an important role in the ubiquitin-proteasome system47. HECTD1 is required for developmental processes in tissues, such as head mesenchyme and neural tube closure48 and placental junctional zone formation49, which are associated with the role of HECTD1 in cell migration50,51. Wnt signaling mediated by APC-Axin interactions is involved in regulating HECTD1 function46. Accordingly, excessive cell migration and proliferation are the obvious feature of the fibrosis process, which has led us to clarify the role of HECTD1 in EndMT-associated fibrosis in patients with silicosis. Moreover, a previous finding of the involvement of another important protein involved in ubiquitination, MCPIP1/ZC3H12A, strongly suggests a role for HECTD1 in silicosis1. The current study has suggested a negative regulatory role for HECTD1 in the EndMT through its ability to inhibit cell viability and migration, consistent with a previous finding showing that HECTD1 inhibits cell migration via Hsp9050. However, the downstream effects of HECTD1 after SiO2 exposure deserve further investigation. Additionally, our analyses of HECTD1 expression in patients with silicosis showed that HECTD1 expression was decreased, consistent with our in vitro results, thus confirming the clinical significance of our findings and revealing that HECTD1 may serve as a potential marker of silicosis.
Conclusion
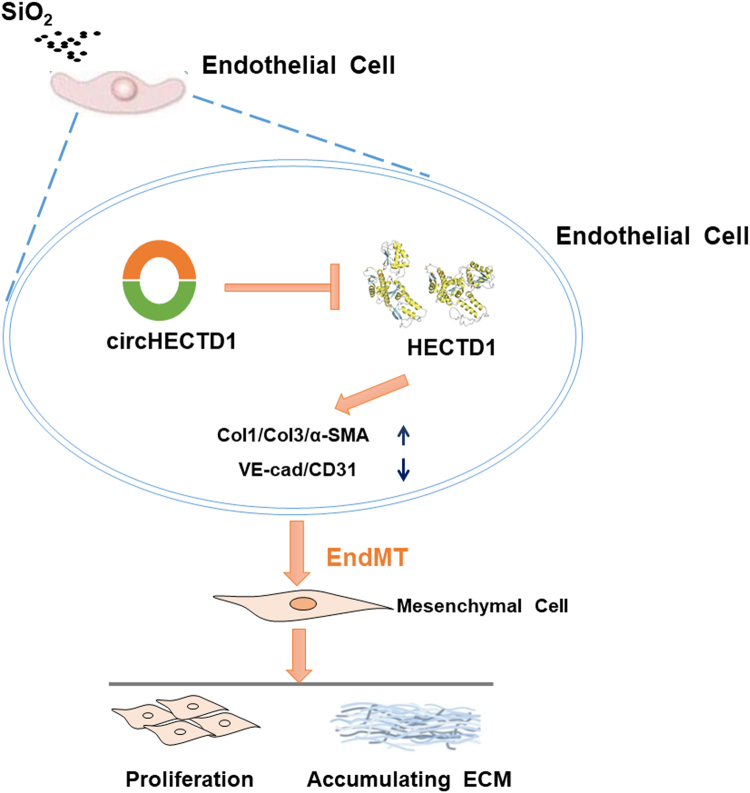
Our study has elucidated a link between the SiO2-induced EndMT and the circHECTD1/HECTD1 pathway, thereby providing insights into the potential use of HECTD1 for developing novel therapeutic strategies for silicosis (Fig. 10).
Fig. 10. Schematic diagram showing the mechanisms by which circHECTD1/HECTD1 regulate the SiO2-induced EndMT.
circHECTD1 expression is increased in endothelial cells exposed to SiO2. The increased expression leads to a subsequent decrease in HECTD1 expression, which, in turn, increases the levels of mesenchymal markers and decreases the levels of endothelial markers. Endothelial cell proliferation and migration are triggered, thereby contributing to irreversible fibrosis in patients with silicosis
Availability of data and materials
All relevant raw data and materials are freely available to any scientist upon request.
Electronic supplementary material
Acknowledgements
This study is the result of work that was partially supported by the resources and facilities of the core laboratory at the Medical School of Southeast University. The National Natural Science Foundation of China (Nos. 81773796, 81473263 and 81600045) and the National Key R&D Program of China (2017YFA0104303).
Authors’ contributions
S.F., H.G., and Y.C. performed the experiments, interpreted the data, prepared the figures, and wrote the manuscript. Z.Z., W.Z., B.H., W.L., and J.W. performed the experiments and interpreted the data. W.X. designed the experiments, interpreted the data, and wrote the manuscript. J.C. provided laboratory space and funding, designed the experiments, interpreted the data, wrote the manuscript, and directed the project. All authors read, discussed, and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All participants provided written informed consent prior to participating in the study. The primary alveolar macrophages derived from human BALF were used in accordance with the approved guidelines from the Research and Development Committee of Nanjing Chest Hospital (2016-KL002-01), and all procedures were conducted in accordance with the Declaration of Helsinki.
Footnotes
These authors contributed equally: Shencun Fang, Huifang Guo, Yusi Cheng.
Edited by A. Oberst.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0432-1).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weiping Xie, Email: wpxie@njmu.edu.cn.
Jie Chao, Phone: +86 25 83272312, Email: chaojie@seu.edu.cn.
References
- 1.Chao J, et al. Role of MCPIP1 in the endothelial–mesenchymal transition induced by silica. Cell. Physiol. Biochem. 2016;40:309–325. doi: 10.1159/000452547. [DOI] [PubMed] [Google Scholar]
- 2.Leung CC, Yu IT, Chen W. Silicosis. Lancet. 2012;379:2008–2018. doi: 10.1016/S0140-6736(12)60235-9. [DOI] [PubMed] [Google Scholar]
- 3.Arciniegas E, Neves CY, Carrillo LM, Zambrano EA, Ramirez R. Endothelial-mesenchymal transition occurs during embryonic pulmonary artery development. Endothelium. 2005;12:193–200. doi: 10.1080/10623320500227283. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto N, et al. Endothelial–mesenchymal transition in bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2010;43:161–172. doi: 10.1165/rcmb.2009-0031OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, et al. Protective effect of spironolactone on endothelial-to-mesenchymal transition in HUVECs via Notch pathway. Cell. Physiol. Biochem. 2015;36:191–200. doi: 10.1159/000374063. [DOI] [PubMed] [Google Scholar]
- 6.Zeisberg EM, Potenta SE, Sugimoto H, Zeisberg M, Kalluri R. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J. Am. Soc. Nephrol. 2008;19:2282–2287. doi: 10.1681/ASN.2008050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 8.Widyantoro B, et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation. 2010;121:2407–2418. doi: 10.1161/CIRCULATIONAHA.110.938217. [DOI] [PubMed] [Google Scholar]
- 9.Piera-Velazquez S, Li Z, Jimenez SA. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am. J. Pathol. 2011;179:1074–1080. doi: 10.1016/j.ajpath.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correia AC, Moonen JR, Brinker MG, Krenning G. FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-beta signaling. J. Cell. Sci. 2016;129:569–579. doi: 10.1242/jcs.176248. [DOI] [PubMed] [Google Scholar]
- 11.Feng B, et al. miR-200b mediates endothelial-to-mesenchymal transition in diabetic cardiomyopathy. Diabetes. 2016;65:768–779. doi: 10.2337/db15-1033. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki HI, et al. Regulation of TGF-beta-mediated endothelial–mesenchymal transition by microRNA-27. J. Biochem. 2017;161:417–420. doi: 10.1093/jb/mvx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J. Biol. Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 15.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Huang R, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, et al. Macrophage-derived MCPIP1 mediates silica-induced pulmonary fibrosis via autophagy. Part. Fibre Toxicol. 2016;13:55. doi: 10.1186/s12989-016-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu T, et al. The role of MCPIP1 in ischemia/reperfusion injury-induced HUVEC migration and apoptosis. Cell. Physiol. Biochem. 2015;37:577–591. doi: 10.1159/000430378. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, et al. BBC3 in macrophages promoted pulmonary fibrosis development through inducing autophagy during silicosis. Cell Death Dis. 2017;8:e2657–e2657. doi: 10.1038/cddis.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, et al. p53/PUMA expression in human pulmonary fibroblasts mediates cell activation and migration in silicosis. Sci. Rep. 2015;5:16900. doi: 10.1038/srep16900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madri JA, Furthmayr H. Collagen polymorphism in the lung. An immunochemical study of pulmonary fibrosis. Hum. Pathol. 1980;11:353–366. doi: 10.1016/S0046-8177(80)80031-1. [DOI] [PubMed] [Google Scholar]
- 24.Seyer JM, Hutcheson ET, Kang AH. Collagen polymorphism in idiopathic chronic pulmonary fibrosis. J. Clin. Invest. 1976;57:1498–1507. doi: 10.1172/JCI108420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulmer JD, et al. Collagen concentration and rates of synthesis in idiopathic pulmonary fibrosis. Am. Rev. Respir. Dis. 1980;122:289–301. doi: 10.1164/arrd.1980.122.2.289. [DOI] [PubMed] [Google Scholar]
- 26.Bradley KH, Kawanami O, Ferrans VJ, Crystal RG. The fibroblast of human lung alveolar structures: a differentiated cell with a major role in lung structure and function. Methods Cell Biol. 1980;21A:37–64. doi: 10.1016/S0091-679X(08)60757-8. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Du WW, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehx501.P440. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Wang K, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 32.Zhou ZW, et al. circRNA mediates silica-induced macrophage activation via HECTD1/ZC3H12A-dependent ubiquitination. Theranostics. 2018;8:575–592. doi: 10.7150/thno.21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao J, et al. MCPIP1 regulates fibroblast migration in 3-D collagen matrices downstream of MAP kinases and NF-kappaB. J. Invest. Dermatol. 2015;135:2944–2954. doi: 10.1038/jid.2015.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, et al. MCPIP1 mediates silica-induced cell migration in human pulmonary fibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016;310:L121–L132. doi: 10.1152/ajplung.00278.2015. [DOI] [PubMed] [Google Scholar]
- 35.Zhao MM, et al. Therapeutic effect of exogenous bone marrowderived mesenchymal stem cell transplantation on silicosis via paracrine mechanisms in rats. Mol. Med. Rep. 2013;8:741–746. doi: 10.3892/mmr.2013.1580. [DOI] [PubMed] [Google Scholar]
- 36.Xu M, Qing M, Peng D. Silicon dioxide particles deposited in vessels and cartilage of the femoral head. Yonsei Med. J. 2014;55:1447–1449. doi: 10.3349/ymj.2014.55.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piera-Velazquez S, Mendoza FA, Jimenez SA. Endothelial to mesenchymal transition (EndoMT) in the pathogenesis of human fibrotic diseases. J. Clin. Med. 2016;5:45. doi: 10.3390/jcm5040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi SH, et al. A hypoxia-induced vascular endothelial-to-mesenchymal transition in development of radiation-induced pulmonary fibrosis. Clin. Cancer Res. 2015;21:3716–3726. doi: 10.1158/1078-0432.CCR-14-3193. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Hu J, Liu L. MiR-200a modulates TGF-beta1-induced endothelial-to-mesenchymal shift via suppression of GRB2 in HAECs. Biomed. Pharmacother. 2017;95:215–222. doi: 10.1016/j.biopha.2017.07.104. [DOI] [PubMed] [Google Scholar]
- 40.Hou LD, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int. J. Immunopathol. Pharmacol. 2017;30:1–6. doi: 10.1177/0394632016686985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 42.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao Y, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui W, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol. Rep. 2017;37:1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 45.Cao Z, et al. circHIPK2-mediated sigma-1R promotes endoplasmic reticulum stress in human pulmonary fibroblasts exposed to silica. Cell Death Dis. 2017;8:3212. doi: 10.1038/s41419-017-0017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran H, et al. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J. Biol. Chem. 2013;288:3753–3767. doi: 10.1074/jbc.M112.415240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng S, Huang C. E3 ubiquitin ligases in regulating stress fiber, lamellipodium, and focal adhesion dynamics. Cell Adhes. Migr. 2014;8:49–54. doi: 10.4161/cam.27480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zohn IE, Anderson KV, Niswander L. The Hectd1 ubiquitin ligase is required for development of the head mesenchyme and neural tube closure. Dev. Biol. 2007;306:208–221. doi: 10.1016/j.ydbio.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar AA, et al. Hectd1 is required for development of the junctional zone of the placenta. Dev. Biol. 2014;392:368–380. doi: 10.1016/j.ydbio.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkar AA, Zohn IE. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J. Cell Biol. 2012;196:789–800. doi: 10.1083/jcb.201105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, et al. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J. Cell Sci. 2013;126(Pt 12):2617–2628. doi: 10.1242/jcs.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant raw data and materials are freely available to any scientist upon request.