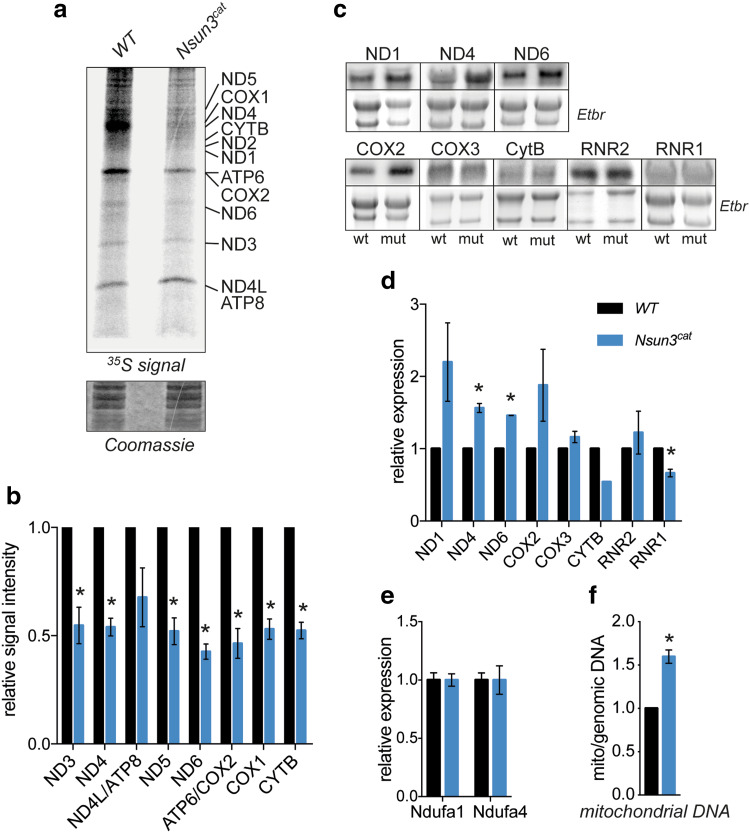
Fig. 3.
Inactivation of Nsun3 compromises mitochondrial protein translation and transcription. a Metabolic labelling experiments show reduced mitochondrial protein translation in Nsun3 cat/cat ESCs. 35S-pulse-labelled mitochondrial proteins were fractionated on a 16% Tricine gel, stained with Coomassie brilliant blue (lower panel), dried, and exposed to phosphoscreen. b Quantification of radiation signals from a for the indicated mitochondrial proteins. Values represent mean ± SEM of three experiments. Statistical significance was calculated by multiple unpaired t test with Holm–Sidak correction (*p < 0.05). c Northern blot analysis of mitochondrial transcripts in wild-type and Nsun3 cat/cat ESCs. Chemiluminescence signals of indicated transcripts (upper panels) and corresponding ethidium bromide (Etbr) stained 28S and 18S rRNA are shown. d Quantification of Northern blot signals shown in c normalized against rRNA band intensities. Values represent mean ± SEM of three experiments except for CytB where only one experiment was quantifiable. Statistical significance was calculated by multiple unpaired t test with Holm–Sidak correction (*p < 0.05). e Expression of nucleus-encoded Ndufa1 and Ndufa4 (electron-chain complex I components) transcripts was analyzed by RT-qPCR. Values represent mean ± SEM of three experiments. Statistical significance was calculated by unpaired t test (*p < 0.05). f Mitochondrial DNA content in Nsun3 cat/cat ESCs was determined by qPCR of a mitochondrial gene normalized against a genomic gene and expressed relative to wild-type ESCs. Values represent mean ± SEM of three experiments. Statistical significance was calculated by unpaired t test (*p < 0.05)