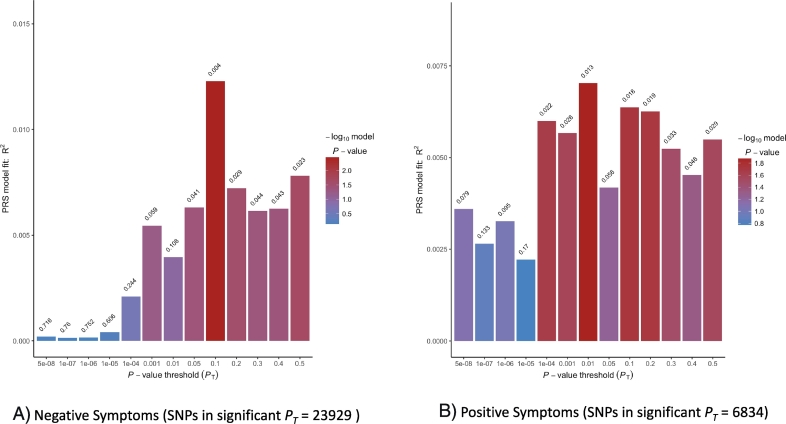
Fig. 2.
Model 2 fit for polygenic risk score predictions on positive and negative symptom dimensions. The plots show model 2 results of 13 analyses based on SNP set without linkage disequilibrium. For each PT, SNPs are selected if significant at that threshold and coefficients and effect sizes estimated. Values above each bar are unadjusted p-values of phenotype from regression analyses. For 2A, the best fit PRS for negative symptoms is at PT of 0.1 and explains roughly 1.2% of the variance. For 2B, the best fit PRS for positive symptoms is at PT of 0.01 and roughly explains 0.7% of the variance.