Figure EV3. Dynamic expression patterns for selected regulators and markers.
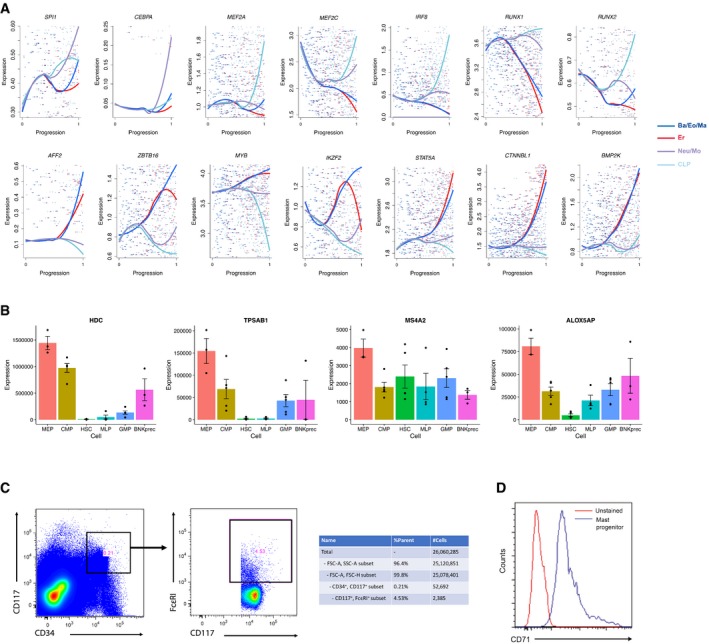
- Shown are the expression levels of canonical and novel regulators across the four trajectories, using the same color scheme as Fig 3B. X‐axis reflects the developmental progression across each trajectory, representing the normalized distance from the MST root (Materials and Methods). A local polynomial regression with span = 0.9 was used for smoothing, with the underlying gene expression data shown as individual points.
- The normalized expression for four Ba/Eo/Ma markers, taken from a microarray dataset of reference progenitor populations (Laurenti et al, 2013). Error bars indicate standard errors across biological replicates (three replicates per population) from the original dataset. For all of these markers, expression is highest in the MEP population, indicating that our Ba/Eo/Ma population is traditionally mixed into the standard MEP gate.
- Gating strategy to enrich for mast cell progenitors (CD34+ CD117+ FcεRIα +) from human umbilical cord blood mononuclear cells, based on the sorting panel from Dahlin et al (2016). Accompanying table shows the proportion of each sorting gate and sorted cell numbers.
- Flow cytometry result showing CD71 protein expression on the surface of mast cell progenitors compared with unstained control, showing the enrichment of CD71 surface expression on mast cell progenitors.
