Fig. 1.
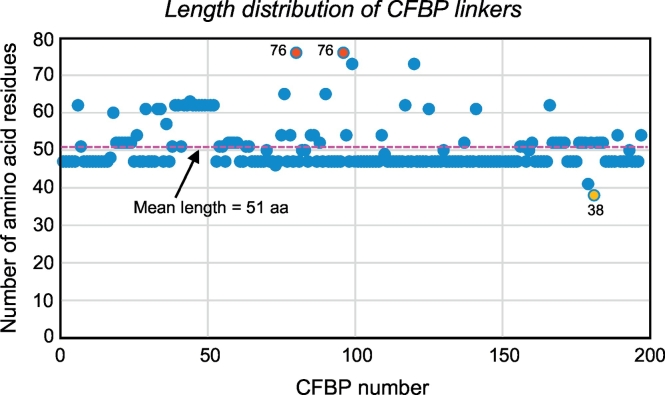
Length distribution of CFBP linkers. The number of amino acid residues in all 277 CFBP linkers was manually counted and tabulated in Excel (not shown). In plotting them in graph, however, the over-crowding was minimized by removing sequences that were in the same phylogenetic clade (from Clustal Omega ordering) and nearly identical to each other (>90% identical), while including many representative ones (Supplementary Material 1). For instance, Treponema putidum CFBP was removed because it was 94% identical to Treponema denticola CFBP. However, the mean length (51 amino acids, pink dotted line in color) was in fact calculated from the full roster of 277 sequences; the shortest (38 amino acids, orange in color) and two longest sequences (76 amino acids, red in color) were also not removed, as they were evidently different from all others. The final graph, shown here, represents 197 sequences. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Length distribution of CFBP linkers. The number of amino acid residues in all 277 CFBP linkers was manually counted and tabulated in Excel (not shown). In plotting them in graph, however, the over-crowding was minimized by removing sequences that were in the same phylogenetic clade (from Clustal Omega ordering) and nearly identical to each other (>90% identical), while including many representative ones (Supplementary Material 1). For instance, Treponema putidum CFBP was removed because it was 94% identical to Treponema denticola CFBP. However, the mean length (51 amino acids, pink dotted line in color) was in fact calculated from the full roster of 277 sequences; the shortest (38 amino acids, orange in color) and two longest sequences (76 amino acids, red in color) were also not removed, as they were evidently different from all others. The final graph, shown here, represents 197 sequences.