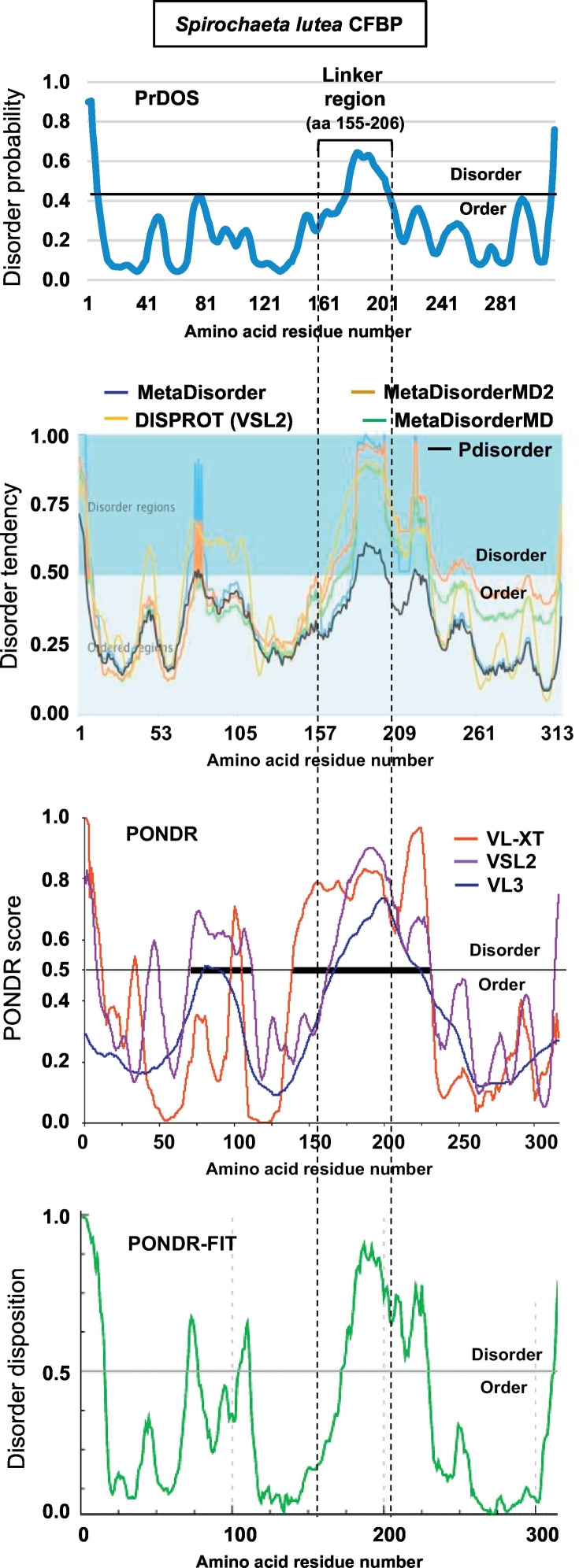
Fig. 3.
Comparison of multiple disorder prediction programs and validation of PrDOS. The Spirochaeta lutea CFBP (with a 52-amino acid linker, close to the average linker size in all CFBPs, see Fig. 1), was arbitrarily chosen for disorder prediction by multiple programs that are freely available, as described in Materials and Methods [17, 18]. The PrDOS result (used in the rest of the paper) is compared with the major programs as shown. The default order/disorder cut-off for each program is indicated, and 8% FPR (false positive rate) was chosen for PrDOS, which was translated into 0.43 cut-off, as explained in Materials and Methods. The default terminology and scale of the respective programs were also used to describe their Y-axes. The dotted lines running through all plots bracket the linker sequence (amino acid 155–206) and show the agreement among the programs. Note that in this particular CFBP, the disorder happens to be slanted towards the C-terminal half of the linker. The high spike of disorder in the two termini of the CFBP is a general feature of most proteins, and is to be ignored for this study.