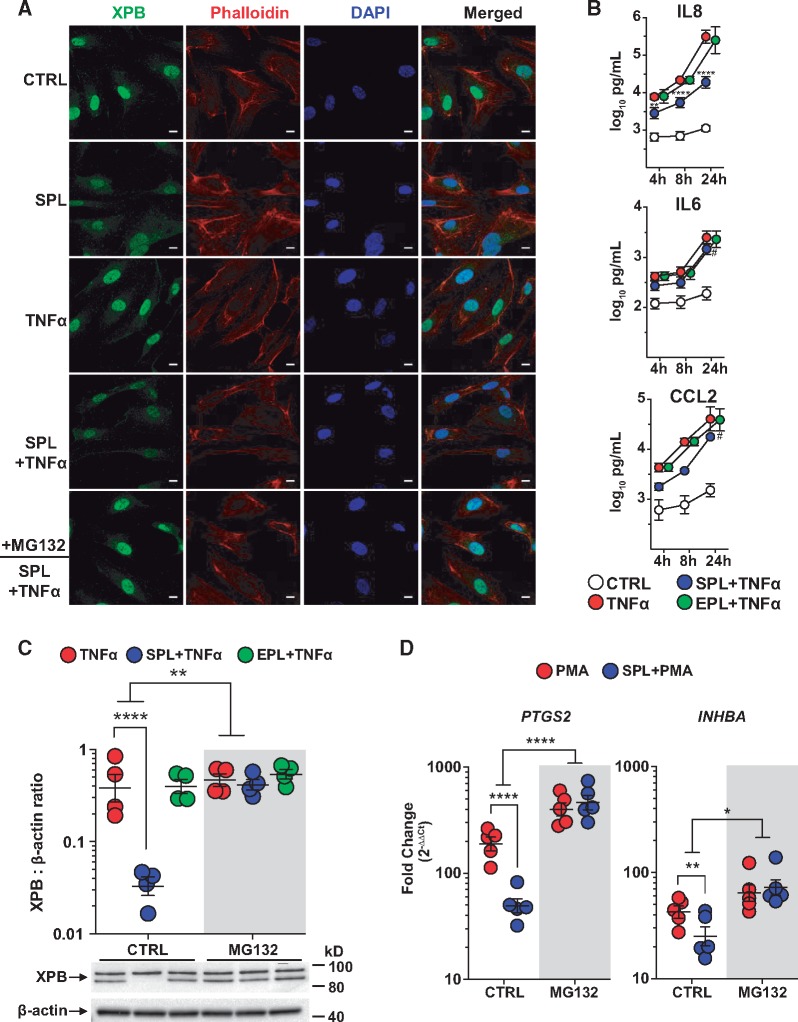
Figure 3.
SPL suppression of inflammation in PAECs and the role of XPB degradation. (A) XPB protein expression, localized predominantly to the nucleus, was reduced following treatment with SPL. The effect of SPL on XPB (green) appeared similar in the absence or presence of TNFα stimulation for 2 h and was blocked by proteasome inhibition (MG132; 10 µM). Cell cytoplasm was stained with phalloidin (red) and nuclei were counterstained with DAPI (blue). Fluorescence images are representative of results from three different PAEC donors. Scale bars: 10 μm. (B) In PAECs, SPL (10 µM) significantly suppressed TNFα-induced secretion of IL8, IL6, and CCL2, while EPL (10 µM) had no effect on any of these cytokines (P ≥ 0.77 for the EPL main effect across all three time points). Data are presented as the log10-transformed mean concentration ± SE of four independent experiments using different donors. Multiplex cytokine analysis was performed on cell supernatant collected at 4, 8, and 24 h after TNFα (5 ng/ml) stimulation. **P < 0.01; ****P < 0.0001 for the effect of SPL at 4, 8, and 24 h, respectively; #P ≤ 0.0002 for the SPL main effect, when similar across all three time points. See Supplementary material online, Table S1 for the results of all cytokines analysed. Consistent with findings by immunofluorescence, (C) MG132 blocked SPL-induced XPB degradation in PAECs as determined by total cell lysate Western blots. In contrast to SPL, EPL had no effect on XPB protein levels in the absence or presence of MG132 (P ≥ 0.30 for both). Densitometric quantification of XPB protein expression relative to β-actin is presented as the geometric mean ratio ± geometric SE on log10 scale of four independent experiments using different donors. A representative Western blot is shown below the graph. (D) MG132 significantly blocked the effect of SPL on PMA-induced PTGS2 and INHBA mRNA expression. PAECs were treated with SPL for 1 h followed by stimulation with PMA (10 nM) for 4 h. Expression of mRNA measured by quantitative real-time PCR is presented as fold-change relative to unstimulated cells (geometric mean ± geometric SE) of five independent experiments, each with a different donor, plotted on log10 scale. *P < 0.05; **P < 0.01; ****P < 0.0001 (ANOVA with post hoc tests, as indicated).