Fig. 4.
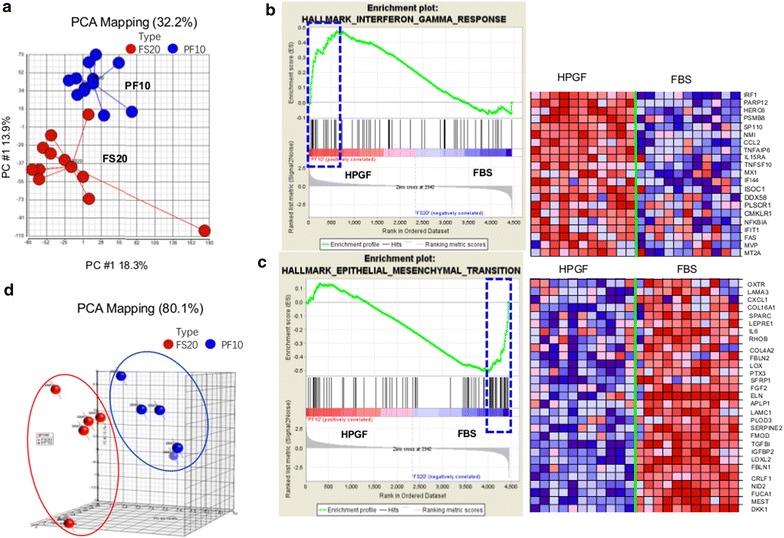
Global transcriptome and microRNA analysis of BMSCs cultured in 10% HPGF-C18 and 20% FBS. BMSCs were prepared from marrow aspirates of 5 healthy subjects and were cultured for 4 passages in 10% HPGF-C18 or 20% FBS. Passage 3 and 4 BMSCs from all 5 subjects were analyzed by global gene expression analysis. The results of the principal component analysis (PCA) analysis of the gene expression data are shown, the blue circles represent BMSCs cultured in 10% HPGF-C18 and the red circles BMSCs cultured in 20% FBS (a). Microarray data were analyzed using Gene Set Enrichment Analysis (GSEA) software to identify functionally related groups of genes (gene sets) with statistically significant enrichment. The enrichment plot for interferon gamma response and the 21 genes on the leading edge that are positively correlated with 10% HPGF-C18 (indicated by the blue rectangle) are shown in b. The enrichment plot for epithelial–mesenchymal transition and the 29 genes on the leading edge that are negatively correlated with HPGF-C18 culture (as indicated by the blue rectangle) were shown in c. In both b, c the plot on the left shows the distribution of genes in the set that are positively and negatively correlated with the HPGF-C18 phenotype. The plot on the right shows the relative gene expression (red = high, blue = low) for each gene for the indicated samples. Passage 4 BMCSs were also analyzed by microRNA PCR array, the PCA results on the microRNAs were shown, the blue circles represent BMSCs cultured in 10% HPGF-C18 and the red circles BMSCs cultured in 20% FBS (d)
