Overexpression of SVP2 in kiwifruit delays budbreak before sufficient winter chilling. SVP2-mediated vegetative growth restriction involves stress response pathways, and commonalities exist between Arabidopsis and kiwifruit SVP targets.
Keywords: ABA, Actinidia, budbreak, bud dormancy, dehydration, kiwifruit, SVP, transcriptome
Abstract
MADS-box genes similar to Arabidopsis thaliana SHORT VEGETATIVE PHASE (SVP) have been implicated in regulation of flowering in annual species and winter dormancy in perennial species. However, the underlying regulatory mechanisms remain to be identified. In this study, the role of kiwifruit SVP2 was explored using ectopic transgenic expression in kiwifruit species with different chilling requirements and the model species tobacco, followed by transcriptomic analysis of transgenic kiwifruit plants. Ectopic expression of SVP2 affected the duration of dormancy in a high-chill kiwifruit Actinidia deliciosa. This effect could be overcome by sufficient winter chilling. SVP2 had a minimal effect on the duration of dormancy in a low-chill kiwifruit A. eriantha. Expression in a tobacco cultivar with photoperiodic regulation of flowering resulted in retarded vegetative growth but no impact on flowering. Transcriptomic analyses of the kiwifruit SVP2 transgenic and control lines identified 92 significantly differentially expressed genes potentially involved in SVP2-mediated growth repression during dormancy, suggesting a role complementary to abscisic acid (ABA). This study has demonstrated that kiwifruit SVP2 has an integrative role in suppression of meristem activity to prevent precocious budbreak before the fulfilment of winter chilling requirements.
Introduction
In temperate horticultural woody perennials, winter dormancy is of particular importance both to avoid unfavourable winter conditions, and to synchronize budbreak and flowering in the following spring (Cooke et al., 2012; Yamane, 2014). Winter dormancy is a dynamic process, defined as a period between bud set in the autumn and budbreak in the spring, when no visible growth occurs. Dormancy has been divided into para-, endo-, and eco-dormancy phases (Lang et al., 1987). Para-dormancy is the suspension of growth caused by factors outside the meristem but within the plant, such as apical dominance. Endo-dormancy is the deepest state of dormancy, when budbreak is prevented by endogenous factors specific to the meristem, which stop the growth even under favourable external conditions. Eco-dormancy is when the growth capacity is restored in the meristem, but remains suspended because of unfavourable external environmental factors and can be released when conditions become permissive. A certain amount of chilling in a bud is often required for the transition from endo-dormancy to eco-dormancy (Lang et al., 1987; Rohde et al., 2007).
Bud development can be dissected into bud formation, acclimation to dehydration and cold, and dormancy. Each of these steps is associated with specific sets of regulatory and marker genes and metabolites (Ruttink et al., 2007). Recent studies of metabolites and gene expression reconstruct the temporal sequence of events during bud development. At least three main regulatory programmes control the onset of dormancy, namely signal perception, hormone alteration, and transcription factors (Shim et al., 2014). Similarly, multiple functional categories of differentially expressed genes (DEGs) have been identified during dormancy release, including stress response, sugar metabolism, hormone response, cell cycle and DNA processing, energy generation, transcription factors, and signal transduction (Fabbroni, 2009). This has been further reinforced by a number of independent studies (Walton et al., 2009; Leida et al., 2012; Liu et al., 2012; Nishitani et al., 2012; Bai et al., 2013; da Silveira Falavigna et al., 2013; Ueno et al., 2013; Zhong et al., 2013; Howe et al., 2015). However, genetic regulation of dormancy remains largely unknown.
The first suggestion that MADS-box genes might be important regulators of dormancy came from a study of the peach (Prunus persica) evergrowing (evg) mutant. Deletion of six tandem arrayed DORMANCY-ASSOCIATED MADS-BOX (DAM) genes in peach resulted in a complete lack of dormancy under cold or short-day (SD) induction, while the expression of a subset of these genes was elevated during endo-dormancy (Bielenberg et al., 2008; Li et al., 2009; Yamane et al., 2011). Similarly, a negative correlation of expression with endo-dormancy release was observed for six tandem arrayed DAM genes predicted to act as transcriptional repressors in Japanese apricot (Prunus mume) (Yamane et al., 2008; Sasaki et al., 2011). Ectopic expression of one of these genes in transgenic poplar resulted in premature growth cessation and terminal bud set, demonstrating a role in growth inhibition in the model woody perennial plant (Sasaki et al., 2011). Genes encoding homologs of DAM transcription factors are differentially regulated during dormancy in many horticultural woody perennials (Mazzitelli et al., 2007; Bielenberg et al., 2008; Diaz-Riquelme et al., 2009; Li et al., 2010; Ubi et al., 2010; Sasaki et al., 2011; Yamane et al., 2011; Liu et al., 2012; Bai et al., 2013; da Silveira Falavigna et al., 2013; Mimida et al., 2015; Porto et al., 2016), suggesting a conserved role in dormancy, and a major quantitative trait locus (QTL) for chilling requirement and bloom date overlapped the peach genomic regions where DAM genes are located (Zhebentyayeva et al., 2014). However, the underlying mechanism and mode of action remain poorly understood and the genetic evidence from diverse species is limited.
DAM proteins are closely related to Arabidopsis thaliana flowering time regulators SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE 24 (AGL24). Arabidopsis SVP and AGL24 are central regulators in the flowering regulatory network, with high sequence similarity but opposite functions. Their mode of action includes interaction with other proteins, resulting in either repressing or activating complexes that regulate floral transition and maintain floral meristem identity (Michaels et al., 2003; Gregis et al., 2006; Lee et al., 2007, 2014; Liu et al., 2007, 2009; Li et al., 2008), or direct binding to the CArG motifs in floral activators, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Lee et al., 2007; Posé et al., 2013). In a herbaceous perennial leafy spurge (Euphorbia esula), DAM-like and FT-like genes are reciprocally and differentially expressed during winter dormancy transition, implying a similar mechanism in regulation of dormancy (Horvath, 2009).
Woody perennials usually have multiple SVP homologues, resulting from lineage- and species-specific expansions within the SVP/AGL24 MADS-box subfamilies (Wells et al., 2015). Four SVP genes have been identified in the kiwifruit species Actinidia chinensis and A. deliciosa, with differential ability to delay flowering in Arabidopsis. Expression of SVP1, SVP2, and SVP4 was elevated in kiwifruit buds over the winter dormancy period, and the relative transcript abundance was higher in colder regions, suggesting roles in bud dormancy and flowering (Wu et al., 2012). In contrast, SVP3 accumulation in buds did not demonstrate seasonal changes, but ectopic expression caused abnormal flower development, reduced petal pigmentation, and abnormal fruit and seed development, supporting a role in repression of reproductive development (Wu et al., 2014). To understand the mechanism of SVP-mediated regulation of kiwifruit bud dormancy, budbreak, and flowering, SVP2 was ectopically expressed in a high chilling requirement species A. deliciosa, a low chilling requirement species A. eriantha, and in Nicotiana tabacum ‘Maryland Mammoth’. Detailed physiological and transcriptomic analyses of 35S:SVP2 A. deliciosa transgenic lines were performed.
Materials and methods
Plant transformation and growth conditions
SVP2 coding sequence under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Wu et al., 2012) was transformed into Agrobacterium tumefaciens strain EHA105 for transformation into kiwifruit A. deliciosa ‘Hayward’ [A. deliciosa (A. Chev.) C.F. Liang et A.R. Ferguson, also referred to as A. chinensis var. deliciosa (A.Chev.) A. Chev.] and A. eriantha Benth. The same construct was transformed into A. tumefaciens strain GV3101 for transformation into tobacco (N. tabacum ‘Maryland Mammoth’). A reporter gene uidA (GUS) under the control of the CaMV 35S promoter (35S:GUS) in appropriate Agrobacterium strains was used to transform control plants. The transformation procedure for A. deliciosa was previously described (Wang et al., 2006, 2007). Transformation of A. eriantha was according to a previously described protocol (Wang et al., 2006, 2007), with modifications to media composition. The regeneration medium contained half-strength Murashige and Skoog (1/2 MS) agar medium (Murashige and Skoog, 1962), 2 mg l−1 6-benzylaminopurine (BAP), 1 mg l−1 zeatin, 0.2 mg l−1 indole-3-butyric acid (IBA), 300 mg l−1 timentin, and 150 mg l−1 kanamycin. The shoot elongation medium contained 1/2 MS, 0.1 mg l−1 zeatin, 0.5 mg l−1 IBA, 300 mg l−1 timentin, and 50 mg l−1 kanamycin. Once their roots were established, transgenic plants were transferred to soil and grown in a containment glasshouse for 18 months at Plant & Food Research, Auckland, New Zealand. Budbreak time and flowering time for transgenic A. deliciosa and A. eriantha were assessed in the following spring season. Nicotiana tabacum transformation was carried out on young leaf discs excised from in vitro grown shoots (Horsch et al., 1985). Transgenic tobacco plants were grown in a containment glasshouse at 20 °C under SD conditions (8/16 h light/dark). The seeds from these transgenic plants were collected and germinated on 1/2 MS agar medium (Murashige and Skoog, 1962) supplemented with 50 µg ml−1 kanamycin. Following the segregation tests, two homozygous lines were chosen and six T2 generation plants of each line were used for detailed analysis.
For clonal propagation, A. deliciosa 35S:SVP2 Line 1 young shoots were collected and surface sterilized using 25% bleach (containing 1.25% sodium hypochlorite) for 20 min, followed by rinsing with sterile water five times. The nodes with axillary buds were excised and transplanted to MS medium. New shoots initiated from these axillary buds, and subsequently seven clonal plants were generated. Once roots were established, plants were transferred to ambient containment glasshouse conditions over 18 months. To initiate dormancy, plants were maintained in SD conditions for 6 weeks (18 °C, 14 h dark and 10 h light intensity at 300–600 µmol s−1 m−2) and subsequently subjected to 4 weeks of fluctuating temperature conditions (14–20 °C during the day and 4–10 °C at night) with an average 9.5 h day length (maximum light intensity at 1000–2000 µmol s−1 m−2). After 100% leaf drop, lateral buds were collected and the plants were subjected to chilling at 3–7 °C for up to 8 weeks.
Determination of A. deliciosa dormancy status
Dormancy status was determined as described previously (Voogd et al., 2015). Briefly, stem cuttings with a single lateral bud were excised on a regular basis from each plant, the lower ends were immersed in water and maintained at budbreak forcing conditions (20 °C, 14 h photoperiod of white light and 70–80% humidity), and the number of days until visible budbreak was recorded. A minimum of three cuttings for each plant were used.
RNA extraction and expression studies
Total RNA was extracted from kiwifruit tissue as previously described (Chang et al., 1993). Total RNA was isolated from tobacco leaf using the Trizol reagent (Invitrogen). A 5 μg aliquot of total RNA was treated with DNase I (Ambion) and reverse transcribed at 37 °C using the BluePrint® Reagent kit for reverse transcription–PCR (RT–PCR) (TaKaRa) according to the manufacturer’s instructions. Amplification and quantification were carried out using the LightCycler® 480 System and SYBR Green I Master Mix (Roche Diagnostics). Reactions were performed in quadruplicate, and a non-template control was included in each run. Thermal cycling conditions were 95 °C for 5 min, followed by 50 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s, followed by a melting temperature cycle, with constant fluorescence data acquisition from 65 °C to 95 °C. The data were analysed using the ratio of target to reference and calculated with the LightCycler®480 software 1.5 (Roche Diagnostics). The expression was normalized to previously characterized reference genes, kiwifruit Actin (Wu et al., 2012) and tobacco Ntα-Tub1 (Pattanaik et al., 2010). Primer sequences used in this study are listed in Supplementary Table S1 at JXB online.
RNA-seq library construction and sequencing
Bud samples were collected from A. deliciosa SVP2 transgenic and control plants on 28 June 2013, 14 August 2013, and 4 October 2013. Three biological replicates from independent overexpressing SVP2 transgenic and control lines were used, with 8–10 lateral buds per replicate at each stage. Total RNA was extracted from lateral buds as previously described (Chang et al., 1993). RNA samples were treated with RNase-free DNase I (Life Technologies, New Zealand) followed by an RNA cleanup kit (Zymo Research). RNA integrity was measured using the RNA 6000 Nano kit and the 2100 Bioanalyzer instrument (Agilent). The library preparation and sequencing were performed by Macrogen, Korea, using Illumina HiSeq™ 2000. Raw sequence data in fastq format were filtered to remove the adaptors and the low-quality reads, leaving a total of 328 million clean reads for subsequent analysis.
De novo transcriptome assembly
The single-end forward reads from each library were initially mapped to the kiwifruit A. chinensis ‘Hongyang’ reference genome (Huang et al., 2013) using Bowtie2 with default settings (Langmead and Salzberg, 2012). The average mapping rates were low, at ~50% (Supplementary Table S2), reflecting actual sequence differences between the two closely related Actinidia species, as well as the poorly predicted and annotated gene models of the draft reference genome. To increase mapping rates, de novo A. deliciosa transcriptome assembly was carried out using the short-read assembly program, Trinity version (0.0.1) (http://trinityrnaseq.sourceforge.net/). Transcriptome assembly completeness was assessed using cegma_v2.4.010312 (http://korflab.ucdavis.edu/datasets/cegma/). False and duplicated contigs were removed using EvidentialGene VERSION 2013.07.27 (http://arthropods.eugenes.org/EvidentialGene/). This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession no. GEYI00000000. A total of 68 454 unique contigs were assembled and annotated using reciprocal BLAST alignment to TAIR 10 Arabidopsis protein databases (https://www.arabidopsis.org). To enrich the reference transcript library, we combined de novo assembled contigs with the A. deliciosa EST library containing 6454 ESTs (Crowhurst et al., 2008) as the reference transcriptome database. The single-end forward reads from each library were mapped to this reference using Bowtie2 (Langmead and Salzberg, 2012). The average mapping rates increased to 75% (Supplementary Table S2) and this set was used for subsequent identification of DEGs. Principal component analysis (PCA) (Stacklies et al., 2007) was performed using the DESeq v2.10 package.
Analysis of differentially expressed genes
The DEGs between each sample set were detected with DESeq v2.10 (Anders and Huber, 2010). The cut off of Padj<0.05 value, followed by the absolute value of logFC (log2 fold change) of not less than 1.0 were considered as significantly differentially expressed genes. The gene expression unit was calculated using the RPKM method (reads per kilobase of transcript per million mapped reads). Annotations were obtained by BLAST of amino acid sequence to Arabidopsis amino acid sequences. Hierarchical clustering as well as heatmap analysis of DEGs were described in McAtee (2014). The best Arabidopsis (TAIR 10) hit was used for Gene Ontology (GO) term classification, and significance was established by singular enrichment analysis (SEA) coupled with available background data of Arabidopsis [false discovery rate (FDR) ≤0.05], using AgriGO Version1.2 (Du et al., 2010). The abiotic stress response and hormone response were established using the Arabidopsis eFP Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007).
Results
Overexpression of SVP2 delays budbreak in a high-chill kiwifruit A. deliciosa
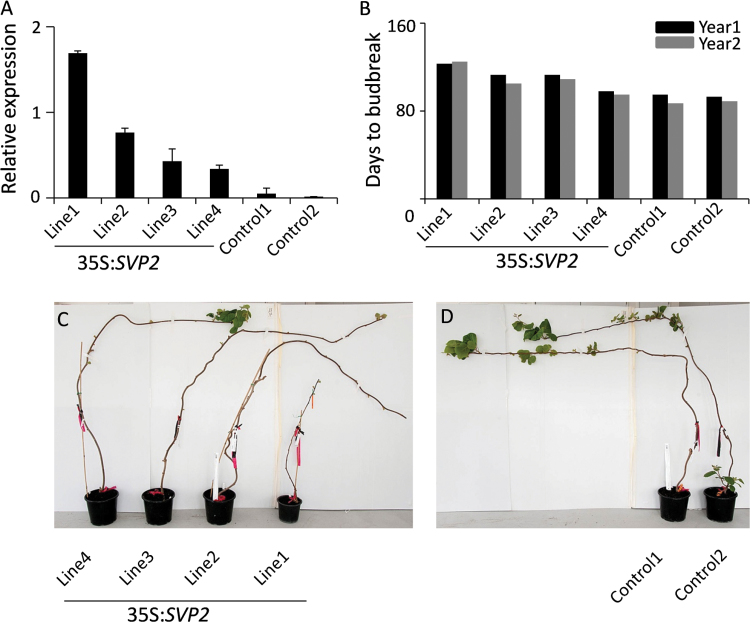
To evaluate the role of SVP2 gene in kiwifruit, transgenic A. deliciosa lines with SVP2 cDNA driven by the CaMV 35S promoter were generated, using the standard transformation protocol (Wang et al., 2012) and the CaMV 35S promoter-driven uidA (GUS) construct as control. Actinidia deliciosa has a high chilling requirement, long dormancy, and late spring budbreak. Normal adventitious shoot formation and growth were observed with the control construct, but the initiation of adventitious buds and shoot elongation were impaired when the SVP2 construct was used. After multiple transformation experiments, four independent transgenic lines were obtained, with varying levels of SVP2 transgene expression (Fig. 1A). No difference in autumn growth cessation, leaf drop, timing of bud-set, and bud formation could be detected between any of the SVP2 transgenic and control lines over the period of 2 years, but a significant delay in the first visible budbreak during the spring season in both years was observed for SVP2 lines. This delay correlated with levels of SVP2 transgene expression and was most prominent in Line 1 (Fig. 1B, C). This line showed slow and weak growth and remained significantly smaller than the control lines over a period of 2 years (Fig. 1C, D).
Fig. 1.
Constitutive expression of SVP2 delays budbreak in Actinidia deliciosa. (A) Relative expression of SVP2 in four 35S:SVP2 transgenic plants and two control plants. The expression was normalized to kiwifruit Actin. Error bars represent the SE of four replicate reactions. (B) Days to budbreak after 100% leaf drop in late autumn. The first visible leaf in spring was recorded as budbreak. (C, D) Transgenic A. deliciosa plants and control plants in the middle of spring.
Cold treatment of SVP2 A. deliciosa transgenic lines
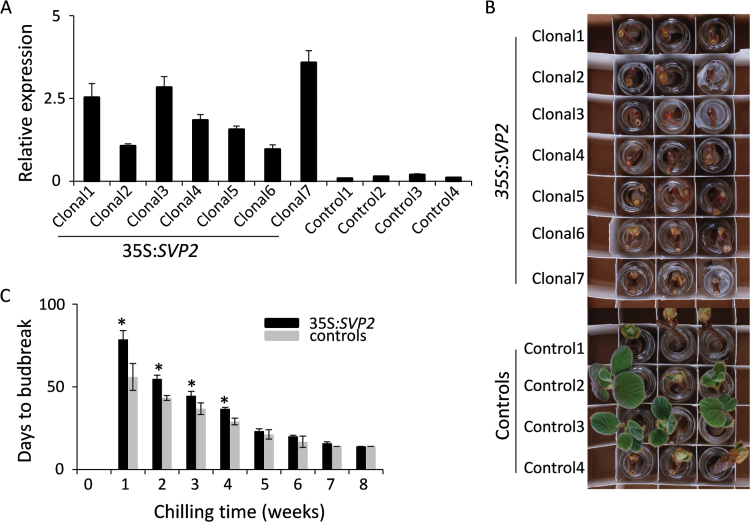
In glasshouse conditions, A. deliciosa plants perceived insufficient chilling because of mild winters in Auckland, New Zealand. To address how chilling conditions related to the SVP2-mediated delay of budbreak, we selected SVP2 Line 1 with the highest transgene expression for further analysis. Seven clonal plants were generated from Line 1, which all expressed the SVP2 transgene (Fig. 2A). The plants were allowed to grow in the containment glasshouse for a period of 18 months, before dormancy was induced by conditions mimicking autumn and early winter. Four control plants were subjected to the same treatment. Dormant plants were exposed to cold treatment for 8 weeks, and three single node cuttings were collected weekly to evaluate dormancy status (Fig. 2B). Average budbreak time demonstrated negative correlation with the duration of chilling (Fig. 2C). Without chilling, no budbreak was observed in cuttings taken from either control or SVP2 plants. In SVP2 plants subjected to cold, a significantly delayed budbreak compared with control plants was detected for up to 4 weeks of cold treatment. A delay of 23, 12, and 8 d was recorded for plants treated for 1, 2, and 3 weeks, respectively. After 4 weeks, average budbreak was still delayed by 8 d, but this delay decreased gradually after 5 weeks of cold treatment to no detectable difference after 8 weeks of cold treatment (Fig. 2C).
Fig. 2.
Chilling alleviates SVP2-mediated repression of budbreak. (A) Relative expression of SVP2 in clonal transgenic and control kiwifruit plants. The expression was normalized to kiwifruit Actin. Error bars represent the SE of four replicate reactions. (B) Delayed budbreak of cuttings from SVP2 transgenic plants after 3 weeks of chilling accumulation. (C) Days to budbreak were calculated as the average date of budbreak of seven clonal transgenic and four control plants. Black and grey bars denote transgenic and control plants, respectively. Error bars represent the SE of budbreak time of seven transgenic and four control plants. Asterisks indicate significantly delayed budbreak time of SVP2 plants versus controls (P<0.05; Student’s t-test).
Overexpression of SVP2 in a low-chill kiwifruit A. eriantha
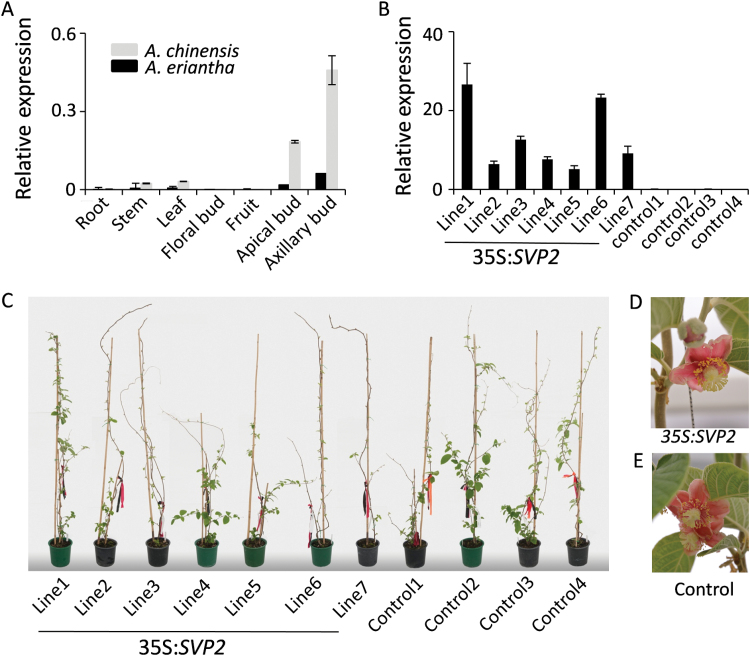
The restored budbreak timing of sufficiently chilled 35S:SVP2 transgenic A. deliciosa prompted us to study the role of SVP2 in a kiwifruit which has a low chilling requirement, A. eriantha. In this kiwifruit species, SVP gene sequences and expression are highly comparable and therefore likely to be functionally conserved as previously described in A. chinensis and A. deliciosa (Wu et al., 2012, 2014) (Fig. 3A). An additional advantage of A. eriantha is the fast reproductive maturity and prolific flowering in glasshouse conditions (Wang et al., 2006), facilitating the study of the role of SVP2 in reproductive onset and development. Initial attempts at regeneration and transformation using standard protocols optimized for A. eriantha (Wang et al., 2006) were unsuccessful. Transformation with the control construct resulted in normal callus formation, initiation of adventitious buds, and subsequent growth, but browning and aborted shoot tip development were recorded with the SVP2 construct (Supplementary Fig. S1). Similar results were obtained in an attempt to transform another kiwifruit species, A. chinensis, suggesting that overexpression of SVP2 plays a detrimental role in regeneration and growth. In an attempt to alleviate transgene-mediated growth restriction, the media were modified to reduce salt concentration (Han et al., 2010) and to change the balance of plant growth regulators, eventually resulting in efficient regeneration, increased initiation of adventitious buds, and improved bud survival. Seven transgenic lines with moderate to high levels of SVP2 transgene expression and four control lines were generated (Fig. 3B) and monitored for budbreak and flowering. A slight delay in the first visible budbreak in some SVP2 transgenic lines was recorded, but no clear correlation could be made between the transgene expression levels and timing of budbreak (Fig. 3C; Table 1). The flowering time and number of flowers was highly variable (Table 1), but all lines produced flowers with normal morphology and pigmentation (Fig. 3D, E).
Fig. 3.
Constitutive expression of SVP2 in a low-chill kiwifruit Actinidia eriantha. (A) Relative expression of SVP2 in A. eriantha and A. chinensis tissues. The level of expression was normalized to Actin. Error bars represent SEs for three replicate reactions. (B) Relative expression of SVP2 in seven 35S:SVP2 transgenic plants and four control plants. The expression was normalized to kiwifruit Actin. Error bars represent the SE of four replicate reactions. (C) Transgenic A. eriantha plants and control plants in early spring. (D, E) Transgenic and control A. eriantha flowers.
Table 1.
Phenotypic analysis of 35S:SVP2 transgenic Actinidia eriantha
Days to budbreak were recorded as days from 100% leaf drop to the first visible budbreak. Number of breaking buds was recorded as the total number of developing shoots at the end of spring. Days to flowering were recorded as days from the first visible budbreak to the appearance of the first floral bud. Number of flowers was counted as total flowers per line
| Transgenic lines | Days to budbreak | Number of breaking buds | Days to flowering | Number of flowers |
|---|---|---|---|---|
| Line 1 | 40 | 28 | 18 | 11 |
| Line 2 | 43 | 21 | 18 | 11 |
| Line 3 | 45 | 23 | 14 | 3 |
| Line 4 | 40 | 25 | 18 | 5 |
| Line 5 | 40 | 20 | 25 | 5 |
| Line 6 | 45 | 23 | 25 | 7 |
| Line 7 | 43 | 22 | 25 | 35 |
| Control 1 | 40 | 32 | nil | 0 |
| Control 2 | 40 | 34 | 32 | 1 |
| Control 3 | 40 | 34 | 25 | 3 |
| Control 4 | 40 | 20 | 18 | 22 |
Overexpression of SVP2 affects plant growth and seed germination, but not flowering time and petal colour in transgenic tobacco
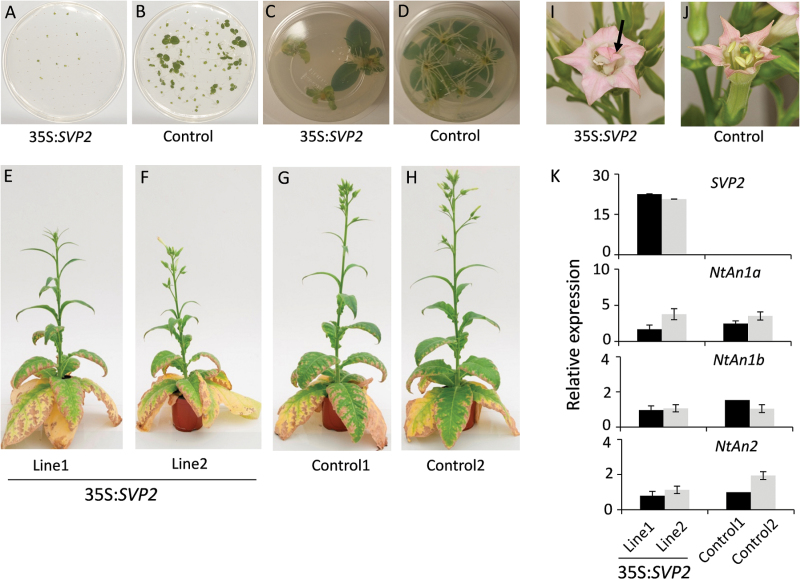
Less efficient transformation in all tested kiwifruit species and delayed budbreak in A. deliciosa suggested that SVP2 played a detrimental role in vegetative growth that could be overcome by changes in growth conditions (chilling and modified media composition), while not affecting flower development and petal colour, in contrast to reports for kiwifruit gene SVP3 (Wu et al., 2014). To investigate whether kiwifruit SVP2 had a conserved growth-restriction effect and further evaluate a role in flowering time and reproductive development, SVP2 cDNA driven by the CaMV 35S promoter was transformed into an SD flowering tobacco variety ‘Maryland Mammoth’. The progeny of two independent transgenic tobacco lines were subjected to detailed analysis. Seed germination and root growth were delayed compared with those in controls (Fig. 4A–D; Table 2). Significant differences were observed in the height of transgenic SVP2 plants, although the plant architecture and secondary growth were visually similar (Fig. 4E–H; Table 2). Flowering time and the number of flowers produced were similar, but more sterile flowers were found on SVP2 plants than on controls (Table 2). Occasional homeotic conversion of stamen to petal was observed (Fig. 4I, J); however, the petal pigmentation and expression of the tobacco anthocyanin regulators, bHLH genes, NtAn1a and NtAn1b, and R2R3 MYB, NtAN2 (Pattanaik et al., 2010; Bai et al., 2011) were comparable with those in control lines (Fig. 4K).
Fig. 4.
Constitutive expression of SVP2 affects vegetative development in transgenic tobacco ‘Maryland Mammoth’. (A, B) Seed germination of 35S:SVP2 plants compared with control plants, 25 d after seeds stratification on MS plates. (C, D) Slow root formation in transgenic 35S:SVP2 plants compared with control plants. (E–H) Two lines of transgenic 35S:SVP2 plants compared with control plants under SD conditions. (I, J) Mutant transgenic SVP2 flower compared with control. The arrow indicates the petaloid stamen in the transgenic flower. (K) Relative expression of NtAn1a, NtAn1b, NtAN2, and the SVP2 transgene in petals of transgenic plants compared with control plants. Black and grey bars represent relative expression of two independent lines. The expression of each gene was normalized to tobacco Ntα-Tub1. Error bars represent the SEs for four replicate reactions.
Table 2.
Phenotypic analysis of 35S:SVP2 transgenic tobacco
Data are presented as means and the SE of six individuals for each lines. Days for seed germination were recorded as days from sterilization to visible germination on MS plates. Total leaf number was counted when the first floral bud was visible. Plant height was expressed as centimetres when the first visible floral bud appeared. Total number of flowers and sterile flowers were counted on inflorescences
| Transgenic Lines | Seed germination (d) | Total leaf number | Plant height (cm) | Total number of flowers | Number of sterile flowers |
|---|---|---|---|---|---|
| Line 1 | 11 ± 2.3 | 18.7 ± 0.4 | 42.5 ± 3.3 | 26.1 ± 1.2 | 21.0 ± 2.5 |
| Line 2 | 13 ± 1.2 | 20.0 ± 1.2 | 31.1 ± 1.1 | 32.5 ± 1.5 | 27.7 ± 4.1 |
| Control 1 | 7 ± 0.0 | 19.3 ± 0.4 | 56.6 ± 3.6 | 28.0 ± 2.5 | 13.5 ± 1.6 |
| Control 2 | 7 ± 0.0 | 22.3 ± 0.4 | 49.0 ± 2.1 | 34.0 ± 2.5 | 14.5 ± 0.5 |
Overexpression of SVP2 in A. deliciosa leads to transcriptomic changes during winter dormancy
To understand the molecular mechanisms underlying SVP2-mediated growth repression in kiwifruit, a transcriptome analysis of SVP2 transgenic lines (t) and control lines (c) at different dormancy stages was performed. Sampling times corresponded to (i) the endo-dormant stage following 100% leaf drop; (ii) transition to eco-dormancy after exposure to winter temperature; and (iii) initiation of budbreak. The dormancy status of bud samples was confirmed by single node cutting assays. Budbreak was delayed in SVP2 plants at the first two time points, and SVP2 plants remained mostly endo-dormant at the first time point (Supplementary Fig. S2). No difference in budbreak time was detected between SVP2 and control lines at the third sampling point. For that reason, only SVP2 transgenic plant bud samples at this time point were analysed further. A total of 15 libraries were prepared and 60–70 million RNA-seq reads were generated for each library. PCA of these RNA-seq reads demonstrated clear separation between sampling dates, but less variation between SVP2 transgenic and control lines at corresponding sampling dates. The sample set at time point 3 was more variable, reflecting transcriptomic changes at an advanced developmental phase just before visible budbreak (Supplementary Fig. S2).
Comparison of SVP2 transgenic and control plant transcriptomes identified 253 genes significantly differentially expressed in the buds collected at the first time point (t1-c1) (Supplementary Table S3), and 226 in the buds collected at the second time point (t2-c2) (Supplementary Table S4), with 92 in the common set, 54 and 38 consistently up- and down-regulated, respectively, in SVP2 transgenic plants (Fig. 5A; Supplementary Table S5). Annotation of the closest Arabidopsis homologue found that 76 DEGs in t1-c1 and 41 DEGs in t2-c2 have been previously identified as direct Arabidopsis SVP targets (Tao et al., 2012; Gregis et al., 2013), while 69 DEGs in t1-c1 and 58 DEGs in t2-c2 have been associated with dormancy in poplar, leafy spurge, and kiwifruit (Horvath et al., 2008; Walton et al., 2009; Howe et al., 2015). Functional classification using GO enrichment analysis identified several categories of biological processes that were significantly affected in the SVP2 transgenic plants, most notably stress response in the t1-c1 and t2-c2 sets. The molecular functions of catalytic and transferase activity were significantly enriched in the common gene set (Supplementary Table S6). Further interrogation of the closest Arabidopsis homologue expression data available through the eFP browser (Winter et al., 2007) revealed that a large proportion of identified genes responded to abiotic stress or plant hormone treatments, most commonly osmotic and cold stress and abscisic acid (ABA) treatment. Of the 92 common genes at both time points, 31 were homologous to Arabidopsis genes that are ABA or osmotic/drought responsive (Supplementary Table S5).
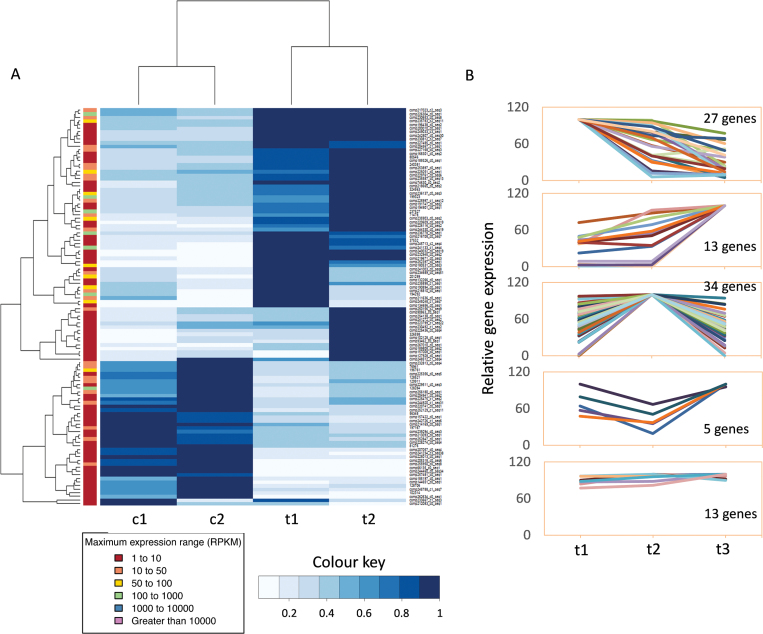
Fig. 5.
SVP2-mediated transcriptomic changes during winter dormancy. (A) Hierarchical clustering of 92 differentially expressed genes identified by transcriptome analysis of SVP2 transgenic lines (t) and control lines (c) at the endo-dormant stage (1) and transition to eco-dormancy after exposure to winter temperature (2). The columns represent comparisons between samples, and rows represent individual genes. The gene name, gene symbol, and RPKM of each gene can be found in Supplementary Table S5 and the higher resolution image is provided as Supplementary Fig. S4. The colour chart of blue and white indicates the RPKM value. Blue and white represent increased and decreased gene expression, respectively. (B) Transcriptome analysis of SVP2 transgenic lines during endo-dormancy (t1), eco-dormancy (t2), and initiation of budbreak (t3) identified five expression patterns for 92 differentially expressed genes.
Analysis of SVP2 plant transcriptomes across different dormancy stages identified five types of expression patterns for the common set of 92 DEGs over the three time points (t1, t2, and t3); increasingly up- or down-regulated from endo-dormancy to budbreak, transiently up- or down-regulated during progression to eco-dormancy, and expressed to a similar level over the dormancy period (Fig. 5B). The accuracy and reproducibility of the transcriptome analysis results was confirmed by real-time RT–PCR analysis of a subset of candidate genes (Supplementary Fig. S3).
Discussion
SVP2 delays shoot outgrowth but may not be sufficient for the onset of dormancy in kiwifruit
In many woody perennials, SVP genes have been associated with winter dormancy. In particular, DAM genes from Prunus persica and P. mume have been advanced as key regulators of winter dormancy (Bielenberg et al., 2008; Jiménez et al., 2010; Sasaki et al., 2011; Yamane et al., 2011). As these Prunus spp. are recalcitrant to transformation, Sasaki et al. (2011) demonstrated a potential role by heterologous expression of Prunus DAM6 in poplar. Overexpression of kiwifruit SVP2 in kiwifruit provides a system to ratify the growth inhibitory function of SVP genes by ectopic expression in the species of origin. The low regeneration efficiency suggested that overexpression of SVP-like genes strongly inhibited outgrowth of plants in tissue culture, potentially explaining the absence of reports on the role of these genes in the species from which they were isolated.
In natural conditions, onset of kiwifruit bud dormancy can be induced by autumn SD and cooler/fluctuating temperature conditions (Brundell, 1976; Lionakis and Schwabe, 1984). The SVP2 kiwifruit lines exhibited no difference in the morphology of the shoot apex and axillary bud formation in comparison with control plants. In particular, premature growth termination and early bud set have not been observed in transgenic A. deliciosa or A. eriantha SVP2 lines grown in ambient conditions in summer (long days). The autumn SD and low temperature conditions did not visibly enhance the leaf senescence, leaf drop, and bud set in transgenic lines. Instead, transgenic plants showed a delay in axillary budbreak in the spring, suggesting that SVP2 was associated with maintenance of deep bud dormancy. This is in contrast to findings reported in transgenic poplar where expression of Prunus DAM6 resulted in premature growth cessation followed by terminal bud set (Sasaki et al., 2011). A possible explanation is that the onsets of terminal and lateral bud dormancy rely on somewhat different mechanisms. In kiwifruit, the shoot tip aborts instead of forming a terminal bud; abortion is preceded by growth cessation and is initiated by tissue necrosis in the subapical zone (Foster et al., 2007). The timing of shoot tip abortion is negatively correlated with the shoot expansion rate and can occur at any time, resulting in short or long shoots. This high developmental plasticity makes visual observations of growth cessation in kiwifruit difficult; however, evidence from both kiwifruit and tobacco SVP2 lines confirms a role in growth inhibition. Delayed germination followed by slower root and shoot development all indicate that SVP2 can act as a growth repressor in tobacco. In addition, both SVP2 and Prunus DAM6 performed a role in lateral bud endo-dormancy in kiwifruit and poplar, respectively (Sasaki et al., 2011), as demonstrated by delayed shoot outgrowth. Therefore, SVP2 in kiwifruit performs as a growth repressor once dormancy has been established, but may not be sufficient to suppress kiwifruit growth in permissive conditions. We therefore propose that SVP2 has a key role in suppressing meristem activity in dormant axillary buds.
Winter chilling can over-ride SVP2-mediated growth inhibition in kiwifruit
Plant dormancy has been divided into three well-defined phases, para-, endo-, and eco-dormancy (Lang et al., 1987). While growth can resume during para- and eco-dormancy, accumulation of chilling is required to release endo-dormancy, to allow budbreak and floral competency in the following spring (Linsley-Noakes and Allan, 1987; Walton et al., 2001; Snelgar et al., 2008). The normal chilling requirement for A. deliciosa ‘Hayward’ is ~800 h (Linsley-Noakes and Allan, 1987), after which dormancy is fully alleviated. Insufficient chilling results in delayed budbreak followed by reduced flower and fruit development. Ectopic SVP2 therefore mimics the effects of insufficient chilling, further delaying budbreak, either by maintenance of deep dormancy or by reduction of shoot outgrowth rate. This effect is gradually reduced and becomes negligible after chilling for the period of ~800 h (5 weeks), suggesting that elevated SVP2 is not sufficient to suppress growth once adequate chilling requirements are met. This finding is consistent with our observations in the low-chill kiwifruit species A. eriantha, where elevated SVP2 had only a minor effect, strongly suggesting that kiwifruit SVP2 does not play a role in chilling-mediated dormancy release. Instead, it would appear that SVP2 prevents premature growth before full chilling is perceived.
Interestingly, elevated expression of SVP2 in shoot buds during dormancy and its decline prior to budbreak (Wu et al., 2012) suggests transcriptional regulation of SVP2 action. This is consistent with other reports of elevated DAM gene expression during dormancy and the suggestions that winter chilling repressed DAM gene expression, resulting in dormancy release (Horvath et al., 2008; Yamane et al., 2008; Li et al., 2009). However, the failure of ectopically expressed SVP2 to maintain dormancy after sufficient chilling indicated additional regulation at the post-transcriptional level. Possible mechanisms are unknown and may include post-transcriptional or post-translational modifications, differential protein stability, or alternative protein–protein interactions. Degradation of SVP protein and differential interactions with other MADS-box protein partners have been established as important during floral transition in Arabidopsis (Michaels et al., 2003; Gregis et al., 2006; Liu et al., 2007, 2009; Lee et al., 2014) and may be instrumental in regulation of dormancy and budbreak in other plant species, including kiwifruit.
SVP2 affects vegetative growth but has no obvious effect on reproductive development and petal colour
Previously, we reported that kiwifruit SVP2 and SVP3 had a differential ability to delay flowering in Arabidopsis and rescue the Arabidopsis svp41 phenotype (Wu et al., 2012). Despite their high sequence similarity, only SVP3 was capable of delaying flowering and complementing the svp41 mutant. Conversely, elevated SVP3 had no obvious effect on vegetative growth, dormancy, or flowering time in transgenic Actinidia or tobacco (Wu et al., 2014), consistent with the lack of increased expression in shoot buds during dormancy (Wu et al., 2012). Instead, elevated SVP3 delayed flower development and reduced petal pigmentation in transgenic A. eriantha and tobacco, through interference with transcription of the key anthocyanin pathway regulators (Wu et al., 2014). While SVP2 had no effect on the timing of flower development and anthocyanin biosynthesis in petals, it was capable of delaying budbreak in the high-chill kiwifruit species and retarding vegetative growth in transgenic tobacco, consistent with elevated expression in shoot buds during dormancy. Therefore, these two closely related kiwifruit SVP genes have acquired different roles as growth repressors in kiwifruit. Co-expression of SVP2 and SVP3 in the shoot buds suggests that they may require different interacting partners to perform diverse functions. Similarly, the inability of the SVP2 transgene to prevent growth in permissive conditions and upon sufficient chilling may indicate a requirement for protein complexes, in which other interacting partners respond directly to environmental stimuli (e.g. accumulation of chilling).
Transcriptomic analysis indicates that SVP genes may mimic the ABA effect
Transcriptomic changes in SVP2 transgenic lines revealed 92 putative SVP2 target genes, significantly up- and down-regulated over two stages of dormancy. Almost half of the genes were typically regulated in response to stress, most often osmotic and cold treatment, with a subset also identified as ABA-responsive genes. These results are consistent with previous findings of coinciding expression of DAM4–DAM6 and several ABA and drought stress response genes during dormancy in peach cultivars (Leida et al., 2012). ABA is an important growth inhibitor previously associated with dormancy; ABA was elevated during endo-dormancy and dropped following the transition to eco-dormancy in several species (Rinne et al., 1994; Le Bris et al., 1999; Rohde and Bhalerao, 2007; Horvath et al., 2008). Consequently, genes associated with response to ABA are often cold, drought, and stress regulated and preferentially expressed during endo- and eco-dormancy (Horvath et al., 2008). ABA affects dormancy progression through its action on dehydrins or membrane permeability (Campoy et al., 2011). Accordingly, kiwifruit genes identified as differentially expressed in SVP2 lines often show homology to well-described genes associated with the dehydration process. Responsive to dehydration 22 (RD22) is a molecular link between ABA signalling and abiotic stress, and its expression has been used as a reliable ABA early response marker in many plants (Shinozaki and Yamaguchi-Shinozaki, 2000; Matus et al., 2014). RD22 has been associated with grape bud dormancy (Mathiason et al., 2009) and Arabidopsis seed dormancy (Yamaguchi-Shinozaki and Shinozaki, 1993). In this current study, two kiwifruit transcripts with homology to RD22 were highly up-regulated at the dormancy stage, but gradually declined prior to budbreak in transgenic SVP2 lines. The Early-Responsive to Dehydration Stress (ERD) genes have been collectively characterized in Arabidopsis as genes that are rapidly induced by dehydration stress. Three transcripts with similarity to an ERD-like gene together with a late embryogenesis abundant protein (ATECP31) were down-regulated in SVP2 overexpression lines, suggesting that different dehydration pathways existed between transgenic and control lines. Other osmotic- and ABA-responsive genes included ASPARAGINASE B1, GLYOXYLASE 17, and Vacuolar processing enzyme, which were all up regulated in SVP2 transgenic lines. Predicted SVP2 targets also include multiple protein kinases and phosphatases, potentially involved in ABA-induced signal transduction, and several transcription factors also associated with stress and ABA.
It is unclear at this stage if ABA metabolism itself is affected by overexpression of SVP2 in kiwifruit. One of the ABA biosynthesis pathway genes, NCED3, was elevated in the SVP2 transgenic lines in the early stage of dormancy (t1-c1), but not differentially expressed at t2-c2. Similarly, we found no evidence of major differences in ABA concentration between transgenic SVP2 and control lines in the samples corresponding to those collected for RNA-seq analysis during dormancy (data not shown). Therefore, it is possible that SVP2 mimics the ABA effect by targeting of genes and pathways associated with the dehydration process. In that case, SVP2 may be targeting the dehydration response only before sufficient chilling is perceived. After sufficient chilling, this pathway may be disrupted and the presence of SVP2 becomes insufficient to repress growth. Interestingly, a significant overlap between kiwifruit SVP2 and Arabidopsis SVP targets was revealed; in particular, in the t1-c1 set, 30% of top Arabidopsis hits have been reported to be directly regulated by SVP (Tao et al., 2012; Gregis et al., 2013), suggesting conservation of the mechanism of action between taxa. However, many of the well-defined Arabidopsis SVP target genes, such as homologues of FT and SOC, were not affected by elevated SVP2 expression, consistent with no demonstrated role for SVP2 in flowering and suggesting that SVP2 preferentially controls only specific aspects of dormancy.
In summary, this study has demonstrated a growth-inhibiting role for kiwifruit SVP2, mediated by ABA and dehydration response pathways, regulating the timing of meristem activity to avoid unfavourable winter conditions and prevent precocious budbreak.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Transformation of Actinidia eriantha.
Fig. S2. Evaluation of A. deliciosa transgenic and control buds at three stages for the RNA-seq experiment.
Fig. S3. qPCR validation of RNA-seq expression profiles.
Fig. S4. High resolution image of hierarchical clustering presented in Fig. 5.
Table S1. qPCR primer sets used in the RNA-seq validation.
Table S2. Summary of RNA-seq experiments.
Table S3. List of the differentially expressed genes (log2 >±1, Padj<0.05) between transgenic SVP2 (t1) and control (c1) in bud samples collected on 28 June 2013.
Table S4. List of the differentially expressed genes (log2 >±1, Padj<0.05) between transgenic SVP2 (t2) and control (c2) in bud samples collected on 14 August 2013.
Table S5. List of the common set of differentially expressed genes in buds collected at both time points (t1-c1 versus t2-c2).
Table S6. List of GO enrichment analysis results (FDR <0.05) for differentially expressed genes in t1-c1, t2-c2, and the common gene set differentially expressed at both time points.
Supplementary Material
Acknowledgements
The authors wish to thank Peter McAtee, Cecilia Deng, and Ross Crowhurst for assistance in bioinformatics analysis, Tim Holmes for photography, Sakuntala Karunairetnam and Andrew Gleave for cloning support, Monica Dragulescu and Wade Wadasinghe for maintenance of plants in the glasshouse, Roger Hellens and Jo Putterill for advise on the project design and progress, and Anne Gunson and Cath Kingston for critical reading of the manuscript. Special thanks to Eric Walton for inspiring our interest in regulation of kiwifruit dormancy and guidance during earlier stages of kiwifruit SVP gene research. This work was funded by the New Zealand Ministry of Business, Innovation & Employment, contract C10X0816 MeriNET. The authors declare no conflict of interest.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Saito T, Sakamoto D, Ito A, Fujii H, Moriguchi T. 2013. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant and Cell Physiology 54, 1132–1151. [DOI] [PubMed] [Google Scholar]
- Bai Y, Pattanaik S, Patra B, Werkman JR, Xie CH, Yuan L. 2011. Flavonoid-related basic helix–loop–helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234, 363–375. [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang Y, Li ZG, Zhebentyayeva T, Fan SH, Reighard GL, Scorza R, Abbott AG. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics and Genomes 4, 495–507. [Google Scholar]
- Brundell DJ. 1976. The effect of chilling on the termination of rest and flower bud development of the Chinese gooseberry. Scientia Horticulturae 4, 175–182. [Google Scholar]
- Campoy J, Ruiz D, Egea J. 2011. Dormancy in temperate fruit trees in a global warming context: a review. Scientia Horticulturae 130, 357–372. [Google Scholar]
- Chang SJ, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116. [Google Scholar]
- Cooke JE, Eriksson ME, Junttila O. 2012. The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant, Cell and Environment 35, 1707–1728. [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. 2008. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira Falavigna V, Porto DD, Buffon V, Margis-Pinheiro M, Pasquali G, Revers LF. 2013. Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Molecular Biology Reporter 32, 796–813. [Google Scholar]
- Díaz-Riquelme J, Lijavetzky D, Martínez-Zapater JM, Carmona MJ. 2009. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiology 149, 354–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbroni C. 2009. Kiwifruit bud release from dormancy: effect of exogenous cytokinins. PhD thesis, The University of Bologna; http://amsdottorato.unibo.it/1966/1/Fabbroni_Cristina.pdf. [Google Scholar]
- Foster TM, Seleznyova AN, Barnett AM. 2007. Independent control of organogenesis and shoot tip abortion are key factors to developmental plasticity in kiwifruit (Actinidia). Annals of Botany 100, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, et al. 2013. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biology 14, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2006. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. The Plant Cell 18, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Gleave AP, Wang T. 2010. Efficient transformation of Actinidia arguta by reducing the strength of basal salts in the medium to alleviate callus browning. Plant Biotechnology Reports 4, 129–138. [Google Scholar]
- Horsch R, Fry J, Hoffmann N, Eichholtz D, Rogers SG, Fraley R. 1985. A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Horvath D. 2009. Common mechanisms regulate flowering and dormancy. Plant Science 177, 523–531. [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. 2008. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GT, Horvath DP, Dharmawardhana P, Priest HD, Mockler TC, Strauss SH. 2015. Extensive transcriptome changes during natural onset and release of vegetative bud dormancy in populus. Frontiers in Plant Science 6, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ding J, Deng D, et al. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nature Communications 4, 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S, Reighard GL, Bielenberg DG. 2010. Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Molecular Biology 73, 157–167. [DOI] [PubMed] [Google Scholar]
- Lang GA, Early JD, Martin GC, Darnell RL. 1987. Endodormancy, paradormancy, and ecodormancy—physiological terminology and classification for dormancy research. Hortscience 22, 371–377. [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bris M, Michaux-Ferrière N, Jacob Y, Poupet A, Barthe P, Guigonis J-M, Le Page-Degivry M-T. 1999. Regulation of bud dormancy by manipulation of ABA in isolated buds of Rosa hybrida cultured in vitro. Functional Plant Biology 26, 273–281. [Google Scholar]
- Lee JH, Chung KS, Kim S-K, Ahn JH. 2014. Post-translational regulation of SHORT VEGETATIVE PHASE as a major mechanism for thermoregulation of flowering. Plant Signaling and Behavior 9, e28193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes and Development 21, 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C, Conesa A, Llácer G, Badenes ML, Ríos G. 2012. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytologist 193, 67–80. [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. 2008. A repressor complex governs the integration of flowering signals in Arabidopsis. Developmental Cell 15, 110–120. [DOI] [PubMed] [Google Scholar]
- Li Z, Reighard GL, Abbott AG, Bielenberg DG. 2009. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) batsch] have distinct seasonal and photoperiodic expression patterns. Journal of Experimental Botany 60, 3521–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZM, Zhang JZ, Mei L, Deng XX, Hu CG, Yao JL. 2010. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Molecular Biology 74, 129–142. [DOI] [PubMed] [Google Scholar]
- Linsley-Noakes G, Allan P. 1987. Effects of winter temperatures on flower development in two clones of kiwifruit Actinidia deliciosa (A. Chev.) CF Liang et AR Ferguson. Scientia Horticulturae 33, 249–260. [Google Scholar]
- Lionakis SM, Schwabe W. 1984. Bud dormancy in the kiwi fruit, Actinidia chinensis planch. Annals of Botany 54, 467–484. [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H. 2009. Regulation of floral patterning by flowering time genes. Developmental Cell 16, 711–722. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. 2007. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910. [DOI] [PubMed] [Google Scholar]
- Liu G, Li W, Zheng P, Xu T, Chen L, Liu D, Hussain S, Teng Y. 2012. Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics 13, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiason K, He D, Grimplet J, Venkateswari J, Galbraith DW, Or E, Fennell A. 2009. Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Functional and Integrative Genomics 9, 81–96. [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Espinoza C, et al. 2014. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS One 9, e110372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S, et al. 2007. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. Journal of Experimental Botany 58, 1035–1045. [DOI] [PubMed] [Google Scholar]
- McAtee PA. 2014. The transcriptional regulation of Actinidia chinensis ‘Hort16A’ fruit ripening. PhD thesis, The University of Auckland; https://researchspace.auckland.ac.nz/handle/2292/23544. [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. 2003. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal 33, 867–874. [DOI] [PubMed] [Google Scholar]
- Mimida N, Saito T, Moriguchi T, Suzuki A, Komori S, Wada M. 2015. Expression of dormancy-associated mads-box (DAM)-like genes in apple. Biologia Plantarum 59, 237–244. [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. [Google Scholar]
- Nishitani C, Saito T, Ubi BE, Shimizu T, Itai A, Saito T, Yamamoto T, Moriguchi T. 2012. Transcriptome analysis of Pyrus pyrifolia leaf buds during transition from endodormancy to ecodormancy. Scientia Horticulturae 147, 49–55. [Google Scholar]
- Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L. 2010. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231, 1061–1076. [DOI] [PubMed] [Google Scholar]
- Porto DD, da Silveira Falavigna V, Arenhart RA, Perini P, Buffon V, Anzanello R, dos Santos HP, Fialho FB, de Oliveira PRD, Revers LF. 2016. Structural genomics and transcriptional characterization of the dormancy-associated MADS-box genes during bud dormancy progression in apple. Tree Genetics and Genomes 12, 1–15. [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. 2013. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. [DOI] [PubMed] [Google Scholar]
- Rinne P, Tuominen H, Junttila O. 1994. Seasonal changes in bud dormancy in relation to bud morphology, water and starch content, and abscisic acid concentration in adult trees of Betula pubescens. Tree Physiology 14, 549–561. [DOI] [PubMed] [Google Scholar]
- Rohde A, Bhalerao RP. 2007. Plant dormancy in the perennial context. Trends in Plant Science 12, 217–223. [DOI] [PubMed] [Google Scholar]
- Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. 2007. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. Journal of Experimental Botany 58, 4047–4060. [DOI] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. 2007. A molecular timetable for apical bud formation and dormancy induction in poplar. The Plant Cell 19, 2370–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R. 2011. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiology 157, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim D, Ko JH, Kim WC, Wang Q, Keathley DE, Han KH. 2014. A molecular framework for seasonal growth–dormancy regulation in perennial plants. Horticulture Research 1, 14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology 3, 217–223. [PubMed] [Google Scholar]
- Snelgar W, Hall A, Mcpherson H. 2008. Modelling flower production of kiwifruit (Actinidia deliciosa) from winter chilling. New Zealand Journal of Crop and Horticultural Science 36, 273–284. [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. 2007. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167. [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. The Plant Journal 70, 549–561. [DOI] [PubMed] [Google Scholar]
- Ubi BE, Sakamoto D, Ban Y, Shimada T, Ito A, Nakajima I, Takemura Y, Tamura F, Saito T, Moriguchi T. 2010. Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. Journal of the American Society for Horticultural Science 135, 174–182. [Google Scholar]
- Ueno S, Klopp C, Leplé JC, Derory J, Noirot C, Léger V, Prince E, Kremer A, Plomion C, Le Provost G. 2013. Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics 14, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd C, Wang T, Varkonyi-Gasic E. 2015. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. Journal of Experimental Botany 66, 4699–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Podivinsky E, Wu RM. 2001. Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiologia Plantarum 111, 396–404. [DOI] [PubMed] [Google Scholar]
- Walton EF, Wu RM, Richardson AC, et al. 2009. A rapid transcriptional activation is induced by the dormancy-breaking chemical hydrogen cyanamide in kiwifruit (Actinidia deliciosa) buds. Journal of Experimental Botany 60, 3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Atkinson R, Janssen B. 2007. Choice of agrobacterium strain for transformation of kiwifruit. Acta Horticulturae 929, 143–148. [Google Scholar]
- Wang T, Karunairetnam S, Wu R, Wang Y-Y, Gleave A. 2012. High efficiency transformation platforms for kiwifruit (Actinidia spp.) functional genomics. Acta Horticulturae 929, 143–148. [Google Scholar]
- Wang T, Ran Y, Atkinson RG, Gleave AP, Cohen D. 2006. Transformation of Actinidia eriantha: a potential species for functional genomics studies in Actinidia. Plant Cell Reports 25, 425–431. [DOI] [PubMed] [Google Scholar]
- Wells CE, Vendramin E, Jimenez Tarodo S, Verde I, Bielenberg DG. 2015. A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biology 15, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wang T, McGie T, Voogd C, Allan AC, Hellens RP, Varkonyi-Gasic E. 2014. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. Journal of Experimental Botany 65, 4985–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RM, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. Journal of Experimental Botany 63, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Molecular and General Genetics 238, 17–25. [DOI] [PubMed] [Google Scholar]
- Yamane H. 2014. Regulation of bud dormancy and bud break in Japanese apricot (Prunus mume Siebold & Zucc.) and peach [Prunus persica (L.) Batsch]: a summary of recent studies. Journal of the Japanese Society for Horticultural Science 83, 187–202. [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. 2008. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. Journal of the American Society for Horticultural Science 133, 708–716. [Google Scholar]
- Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. 2011. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. Journal of Experimental Botany 62, 3481–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhebentyayeva TN, Fan S, Chandra A, Bielenberg DG, Reighard GL, Okie WR, Abbott AG. 2014. Dissection of chilling requirement and bloom date QTLs in peach using a whole genome sequencing of sibling trees from an F2 mapping population. Tree Genetics and Genomes 10, 35–51. [Google Scholar]
- Zhong W, Gao Z, Zhuang W, Shi T, Zhang Z, Ni Z. 2013. Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Molecular Biology 83, 247–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.