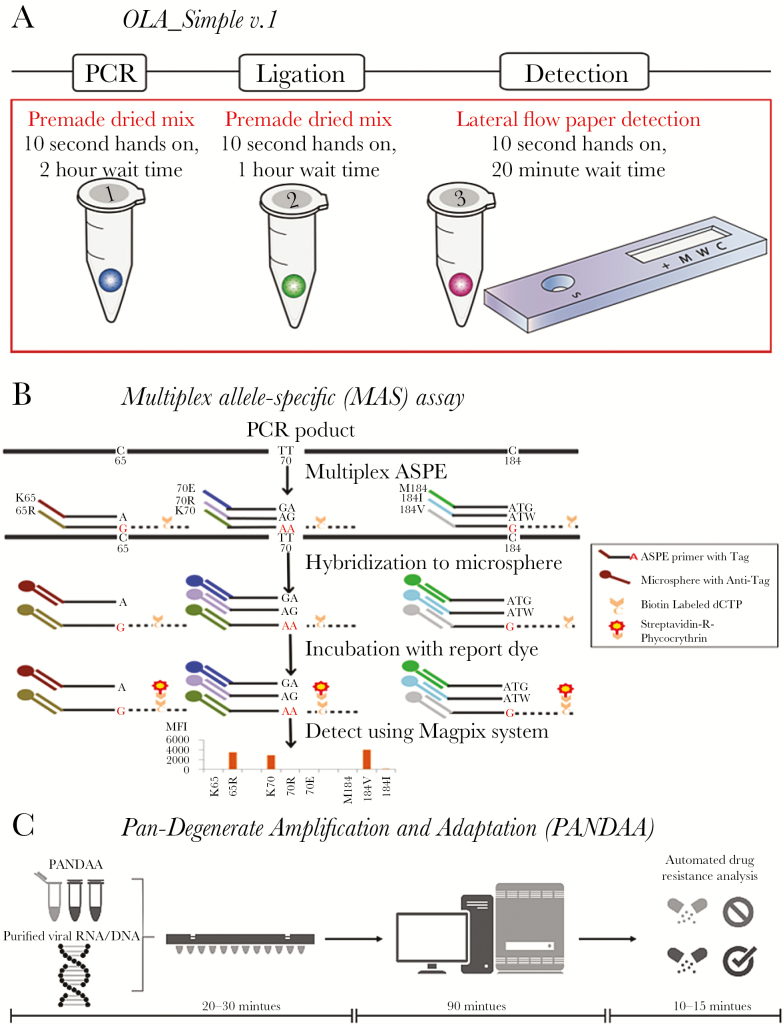
Figure 1.
Point-of-care test to detect human immunodeficiency virus drug resistance. A, Simplified kit with single-use reagents to test one specimen for drug resistance mutations prior to nonnucleoside reverse transcriptase inhibitor–based antiretroviral therapy. Kit detects mutant codons predictive of virologic failure [17]. The kit amplifies DNA using premade dried polymerase chain reaction (PCR) mixture. The product is added to a dried ligation mix and subsequently detected in a paper-strip test cartridge. B, Targets are PCR amplified, then the multiplex allele-specific (MAS) assay uses allele-specific primer extension (ASPE) with specific primers mixed together in one reaction tube containing reaction reagent mixture and a template. When the primer complementary to the 3ʹ-terminal nucleotide of the target, primer extension occurs and biotinylated deoxycytodine triphosphates (dCTPs) are incorporated into the extended products. ASPE products are hybridized to microspheres through the specificity of “TAG”/”Anti-TAG” recognition and read with the suspension array system. C, Premixed pan-degenerate amplification and adaptation (PANDAA) with quantitative PCR (qPCR) enzymes, buffer, primers, and probes labeled with 3 distinct fluorophores to detect 2 drug resistance mutations and quantify total viral nucleic acid. Viral RNA from plasma, DNA from whole blood, or PCR amplicon previously generated for Sanger sequencing can be used as the input template. PANDAA is a one-step reaction that does not require a first-round cDNA synthesis or PCR step prior to qPCR. PANDAA can be run on any qPCR machine that can distinguish the FAM, VIC, and NED fluorophores (or equivalent fluorophores with a similar emission spectra). Automated data analysis allows the relative abundance of each drug resistance mutation to be quantified with additional data handling by the user.