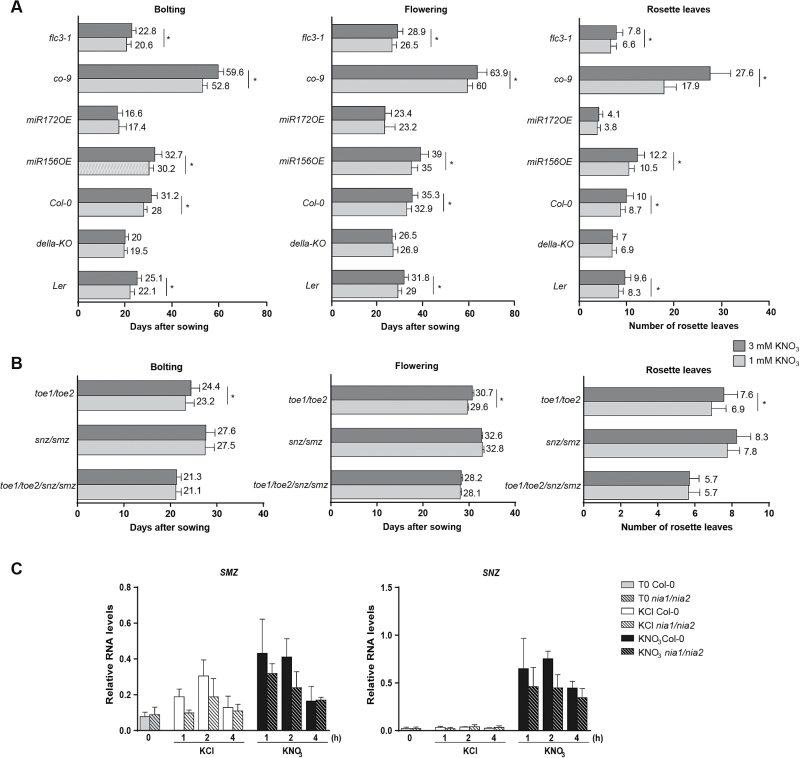
Fig. 2.
Nitrate-dependent delay in flowering time depends on the gibberellin pathway and the miR172 targets SMZ and SNZ. Plants were sown on vermiculite and watered once a week with N-free nutrient solution containing either 1 mM (light gray) or 3 mM (dark gray) KNO3. Nitrate-dependent flowering time and rosette leaf number of mutants for key genes from different flowering pathways were determined (A). Nitrate-dependent flowering and rosette leaf number of the quadruple toe1/toe2/smz/snz or double toe1/toe2 and smz/snz mutants were also quantified (B). (C) Col-0 (filled bars) and nitrate reductase double-mutant (nia1/nia2) plants (dashed bars) were sown on a N-free hydroponic medium supplemented with 0.5 mM ammonium succinate and then treated with either 5 mM KCl or 5 mM KNO3 for the indicated times. Total RNA was extracted from shoots, and SMZ and SNZ transcript levels were quantified by qRT-PCR. The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as an internal reference. flc3-1, FLOWERING LOCUS C mutant; co-9, CONSTANS mutant; miR172OE, miR172 overexpressor; miR156OE, miR156 overexpressor; della-KO, quintuple DELLA mutant (Ler background); toe1, toe2, TARGET OF EAT mutants; snz, SCHNARCHZAPFEN mutant; smz, SCHLAFMÜTZE mutant. All mutants and overexpressor lines, except della-KO, are in the Col-0 background. At least 20 plants were used for each flowering time measurement. Data are means and SD of three independent biological replicates. Asterisks highlight significant differences as determined by Tukey’s Multiple Comparison test (P≤0.01).