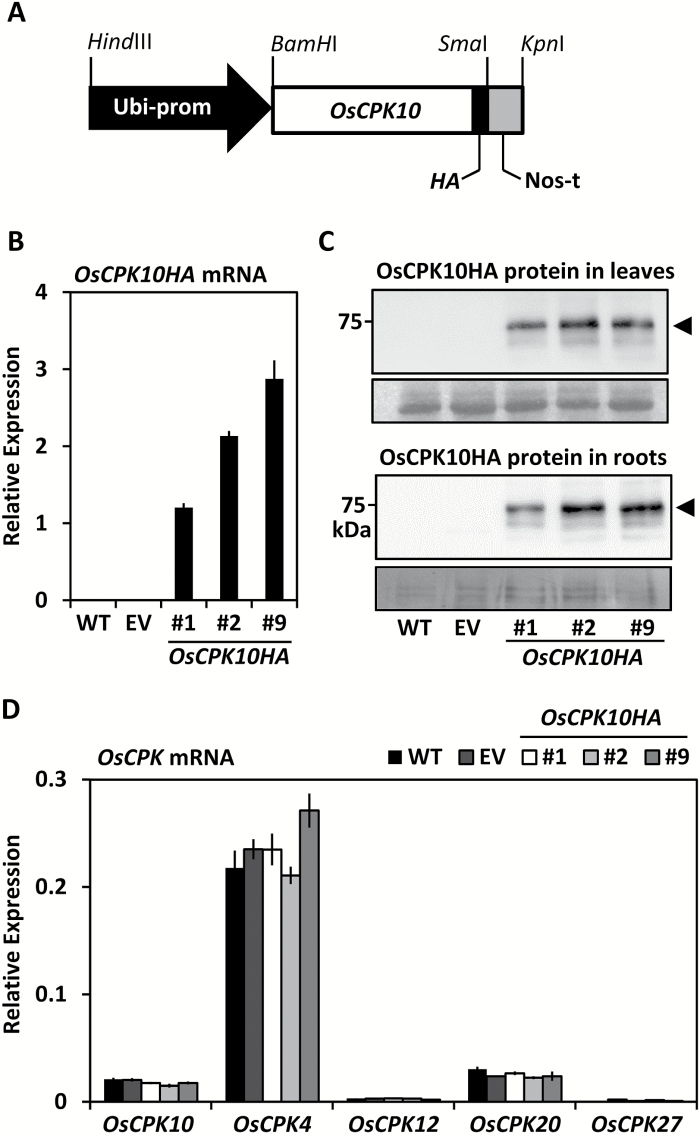
Fig. 2.
OsCPK10HA accumulation in transgenic rice plants. (A) Schematic representation of the pUbi:OsCPK10HA:nos transgene used for rice transformation. (B) Transcript levels of OsCPK10HA in leaves of wild-type (WT), empty vector (EV) and the indicated OsCPK10HA lines as determined by qRT-PCR analysis using OsUbi5 mRNAs for normalization. Values are means and standard deviations of three replicates. (C) OsCPK10HA protein accumulation in leaves and roots as determined by western blot analysis using specific anti-HA antibodies. Lower panels show the loading control for leaves, 25 μg per lane, using Ponceau staining, and for roots, 35 μg per lane, using Coomassie Blue staining. (D) Transcript levels of endogenous OsCPK10 and different OsCPK family members in leaves as determined by qRT-PCR analysis using OsUbi5 mRNAs for normalization. Values are means and standard deviations of three replicates.