Infants hospitalized with RSV A/GA5 bronchiolitis showed greater clinical severity, decreased interferon expression, and enhanced overexpression of neutrophil-related genes, compared with the GA2 or BA genotypes, suggesting the possibility that RSV strain-specific differences may contribute to clinical severity.
Keywords: bronchiolitis, genomic loads, host responses
Abstract
Background
Data on how respiratory syncytial virus (RSV) genotypes influence disease severity and host immune responses is limited. Here, we characterized the genetic variability of RSV during 5 seasons, and evaluated the role of RSV subtypes, genotypes, and viral loads in disease severity and host transcriptional profiles.
Methods
A prospective, observational study was carried out, including a convenience sample of healthy infants hospitalized with RSV bronchiolitis. Nasopharyngeal samples for viral load quantitation, typing, and genotyping, and blood samples for transcriptome analyses were obtained within 24 hours of hospitalization. Multivariate models were constructed to identify virologic and clinical variables predictive of clinical outcomes.
Results
We enrolled 253 infants (median age 2.1 [25%–75% interquartile range] months). RSV A infections predominated over RSV B and showed greater genotype variability. RSV A/GA2, A/GA5, and RSV B/BA were the most common genotypes identified. Compared to GA2 or BA, infants with GA5 infections had higher viral loads. GA5 infections were associated with longer hospital stay, and with less activation of interferon and increased overexpression of neutrophil genes.
Conclusions
RSV A infections were more frequent than RSV B, and displayed greater variability. GA5 infections were associated with enhanced disease severity and distinct host immune responses.
Respiratory syncytial virus (RSV) is the leading cause of hospitalization in infants worldwide. The clinical spectrum of this disease is broad, and varies from a mild upper respiratory illness to severe lower respiratory tract infection
(LRTI) [1].
The role of viral factors such as RSV types and genotypes on clinical parameters of disease severity has been previously evaluated; however, results from different studies are inconclusive (Supplementary Table S1). There are two major RSV types, A and B, which can cocirculate during the same epidemic season. Inconsistent data suggest that RSV A is associated with more severe disease [2–8]. To determine the prevalence of RSV genotypes, molecular epidemiologic studies have been conducted in different parts of the world. In most of those studies, RSV genotyping was performed as part of a respiratory surveillance program in children of all ages, or by analyzing respiratory samples already collected per standard of care. The association between RSV genotypes, specific genotype viral loads, and their influence on clinical disease severity, or the distinct host immune profiles to individual RSV types and genotypes in infants and young children with RSV bronchiolitis have not been characterized in detail [9–17].
The objectives of this study were: (1) to define the variability of RSV A and B genotypes during 5 nonconsecutive respiratory seasons; (2) to determine if RSV subtypes, genotypes, and viral genomic loads related to specific RSV subtypes and genotypes influenced RSV disease severity in young children; and (3) to assess whether host immune profiles differed according to RSV types and genotypes.
MATERIALS AND METHODS
Patient Population and Study Design
Previously healthy infants hospitalized over 5 nonconsecutive respiratory seasons with RSV bronchiolitis at Children’s Medical Center Dallas (CMCD), Texas (2004 to 2009) and Nationwide Children’s Hospital (NCH), Columbus, Ohio (2010 to 2011) were prospectively enrolled. Patients were excluded from the study if they were premature (gestational age ≤36 weeks), had a previous episode of documented RSV infection, chronic medical conditions, immunodeficiency, or had received systemic steroids or immunomodulatory drugs within 2 weeks of hospitalization. Healthy asymptomatic controls were enrolled during well-child visits or minor elective surgical procedures not involving the respiratory tract.
Monday through Friday, infants hospitalized with bronchiolitis were identified using the hospital and microbiology census. Bronchiolitis was defined as the presence of rhinitis, tachypnea, wheezing, cough, crackles, use of accessory muscles, and/or nasal flaring with or without fever [18]. Children who met the inclusion criteria and had a positive RSV test per standard of care, were enrolled within 24 (25%–75% interquartile range [IQR]) hours of hospitalization, and a nasal wash sample was obtained for RSV typing and viral load quantitation by quantitative real time polymerase chain reaction (qrtPCR) following established procedures [19–23]. Nasal wash samples were also tested for other respiratory viruses in 75% of patients; 97% using a multiplex assay targeting 17 pathogens [24] and 3% by viral culture. RNA aliquots were stored at −80°C for further amplification and sequencing as described below. In a subset of infants, blood samples were also collected for transcriptome analysis using microarrays as described [25, 26].
Clinical and demographic data were collected using a clinical questionnaire at the time of enrollment, and by chart review. Clinical illness was assessed using a standardized clinical disease severity score (CDSS) at study enrollment [21, 25], and other parameters of severity including: length of hospital stay, administration and duration of supplemental O2, and pediatric intensive care unit (PICU) admission.
The Institutional Review Boards at the University of Texas Southwestern Medical Center, and Nationwide Children’s Hospital approved the study, classified as a Level 1 risk clinical study, no greater than minimal risk (pursuant under 45 CFR 46.404; and 21 CFR 50.51). Informed consent procedures followed in compliance with Children’s Medical Center, Dallas and Nationwide Children’s Hospital Research Responsible Conduct Guidelines and written informed consent was obtained from guardians before study participation.
Genotype Analyses
Briefly, RNA was extracted from nasal wash samples, and amplification and sequencing of the second hypervariable region of the RSV G gene was performed using protocol A for patients enrolled at CMCD (2004–2009), and protocol B for those enrolled at NCH (2010–2011) (further details are described in the Supplementary Material) [16, 20].
Phylogenetic analysis was performed targeting the C-terminal amino acid sequence of the G glycoprotein based on the methodology described by Peret et al [27]. For samples obtained at CMCD, phylogenetic analysis was performed using MEGA 4.0 [28, 29], and the phylogenetic tree constructed using the neighbor-joining method with bootstraps of 500 replicates. For NCH samples, the phylogenetic analysis was performed using MegAlign of Lasergene 8 program suite (DNASTAR, Inc. Madison, WI). Multiple sequence alignment by the CLUSTAL W and phylogenetic tree methods were obtained comparing the sequences from the study samples with gene sequences of human RSV retrieved from GenBank as references [30]. The genotype of each sequence was assigned according to similarity of sequences to the reference strains as well as the phylogenetic relationship.
Microarray Data Processing and Analyses
Blood samples (1–3 mL) were collected in Tempus tubes (Applied Biosystems, CA) and stored at −20°C within 2–4 hours of collection for further processing in batches. Blood RNA was processed and hybridized into Illumina Human HT12 V4 beadchips (47323 probes) and scanned on the Illumina Beadstation 500, as described [31, 32]. Illumina GenomeStudio software was used for background subtraction and to scale average signal intensities. Briefly, we selected transcripts that were “present” in ≥10% of the samples (PAL10%; 16049 transcripts). Raw expression values below 10 were set to 10 and the data was log2-transformed, as described [25, 26]. For data analyses, we used the limma package in R and applied supervised class comparisons using stringent statistical filters (Benjamini–Hochberg corrected P value < .01, ≥1.25 fold change) to identify transcripts differentially expressed in infants with RSV A or B bronchiolitis, and those infected with GA2, GA5, and BA versus healthy controls [26]. Functional gene analyses were performed using modular repertoires as described [31–33]. This data-driven approach is based on clusters of coordinately expressed genes (modules) that share the same biological function. The list of modular content and annotations is available at http://www.biir.net/public_wikis/module_annotation/V2_Trial_8_Modules. The data are deposited in the NCBI Gene Expression Omnibus (GEO accession number: GSE103842).
Statistical Analysis
We used descriptive statistics to summarize the demographic data and patients’ baseline characteristics. Mann–Whitney or Kruskall–Wallis tests for continuous variables, and the Fisher’s exact or Χ2 test for categorical data were used to compare the different groups. The Benjamini–Hochberg test was applied to correct for multiple comparisons when analyzing families that included multiple parameters (ie, interferon or inflammation modules) and adjusted P values (aP) were reported. We performed multivariable logistic regression analyses to determine if viral factors independently predicted the risk of severe disease defined as length of hospital stay and PICU admission. Firth’s penalized likelihood correction was used for PICU admission to avoid small sample size bias. Variables that were significant in univariate analyses (P < .2) or were biologically meaningful were introduced in the models and included: age (months), gender, clinical disease severity score (which was dichotomized as mild [≤5] versus moderate/severe [>5–15]), RSV genotypes, and viral genomic loads. Associations of predictors with primary outcomes were displayed using relative risk (RR) and 95% confidence intervals (CIs) for length of hospitalization (as continuous outcome) or odds ratios (OR) for PICU admission. Predictor variables with 2-sided P values < .05 and multivariate RR/OR with 95% CIs not including 1 were considered significant. Statistical analyses were performed using SAS 9.3 (SAS Institute Inc, Cary, NC).
RESULTS
Study Subjects
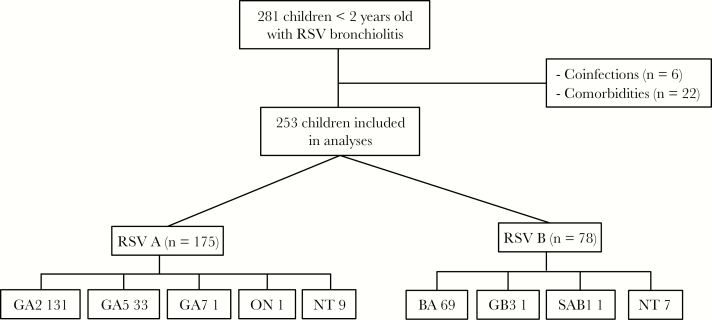
From 1 March 2004 to 30 April 2011, we enrolled 281 infants hospitalized with RSV bronchiolitis. We excluded 28 children from the analyses as outlined in Figure 1: 22 with underlying chronic medical conditions, and 6 with dual RSV A and B infections. The median and IQR age of the remaining 253 children included in further analyses was 2.1 (IQR, 1.1–4.0) months; 57% were males and 62% were white (Table 1).
Figure 1.
Selection of study patients. From March 2004 to April 2011 we enrolled 281 children <2 years of age, hospitalized with respiratory syncytial virus (RSV) bronchiolitis; 28 children were excluded: 6 patients with RSV A and B confections and 22 children with comorbidities including prematurity <36 weeks of gestational age, chronic lung disease, other lung diseases, and congenital heart diseases; 253 children <2 years of age were included in further analyses. Abbreviation: NT: nontyped.
Table 1.
Clinical, Demographic, and Viral Parameters in Infants With Respiratory Syncytial Virus (RSV) A and B Bronchiolitis
| RSV (n = 253) | RSV A (n = 175) | RSV B (n = 78) | P value | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age, months | 2.1 [1.1–4.0] | 2.1 [1.13–4.6] | 2.2 [1.05–3.53] | .71 |
| Gender, n (%) | ||||
| Male | 143 (57) | 101 (58) | 42 (54) | .58 |
| Female | 110 (43) | 74 (42) | 36 (46) | |
| Race/ethnicity, n (%) | ||||
| White | 156 (62) | 115 (66) | 41 (53) | |
| Black | 39 (15) | 28 (16) | 11 (14) | .04 |
| Hispanic | 42 (17) | 25 (14) | 17 (22) | |
| Other | 16 (6) | 7 (4) | 9 (11) | |
| Breastfeeding, % | 38 | 33 | 43 | .22 |
| Second hand smoke, % | 32 | 36 | 28 | .27 |
| Daycare, % | 15 | 19 | 10 | .08 |
| Siblings, % | 84 | 83 | 86 | .58 |
| Disease severity | ||||
| Days of symptoms | 4 [3–6] | 4 [3–6] | 5 [3–7] | .15 |
| O2 administration | ||||
| n (%) | 179 (72) | 127 (73)a | 52 (68)b | .37 |
| Duration, days | 1[0–3] | 1 [0–3] | 1 [0–3] | .86 |
| PICU, n (%) | 26 (10) | 38 (22) | 15 (19) | .73 |
| CDSS | 5 [3–8] | 5 [3–8] | 5 [3–7] | .36 |
| LOS (days) | 3 [2–5] | 3 [2–5] | 3 [2–6] | .48 |
| Viral loads (log10copies/mL) | 5.82 [4.84–6.51] | 6.08 [5.09–6.56] | 5.29 [4.40–8.94] | <.01 |
P values. Continuous variables were analyzed using Mann–Whitney tests and categorical variables using Fisher’s exact or Chi-square test. Data are reported as medians, 25%–75% interquartile range.
Abbreviations: PICU, pediatric intensive care unit; CDSS, clinical disease severity score; LOS, length of stay.
aFor RSV A in 2 patients O2 requirement was not documented.
bFor RSV B O2 requirement was not documented in 1 patient.
Bold highlights the significant.
Patient Characteristics According to RSV Types and Genomic RSV Loads
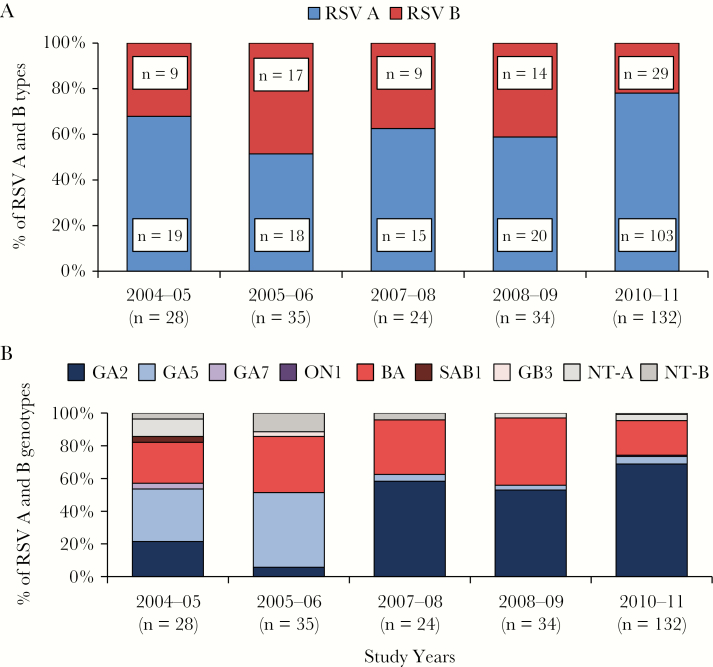
During each of the 5 respiratory seasons studied, there was cocirculation of both RSV types, RSV A and B (Figure 2A); however, RSV A infections were significantly more frequent than RSV B (69% vs 31%; P < .001).
Figure 2.
Distribution of respiratory syncytial virus (RSV) types and RSV genotypes in infants with RSV bronchiolitis from 2004 to 2011. A, The horizontal (x) axis represents the study years and the vertical (y) axis the percentage of RSV A and B isolates. B, The horizontal (x) axis represents the study years and the vertical (y) axis the percentage of RSV A and B genotypes. Abbreviation: NT: nontyped.
Comparing the baseline demographic characteristics and parameters of disease severity between patients with RSV A (n = 175) and RSV B infection (n = 78), there were no significant differences with regards to age or gender, but there were differences in race/ethnicity between groups, likely related to the enrollment site (Table 1). Breastfeeding history, second-hand smoke exposure, attendance to day care, and number of siblings were comparable between groups. Infants infected with RSV A or B presented to the hospital with a median of 4 (IQR 3–6) and 5 (IQR 3–7) days of symptoms, respectively (P = .15).
RSV genomic loads in nasal wash samples ranged from 4.4 to 6.6 log10 RNA copies/mL and were inversely correlated with days of illness before hospital admission (r = −0.22; P < .001). Although viral genomic loads were significantly higher for RSV A versus RSV B infection (6.1 vs 5.3 log10 copies/mL, respectively; P < .01) and independent of duration of illness, we did not observe differences in clinical disease severity between groups as defined by: CDSS, administration and duration of supplemental oxygen, PICU admission, and duration of hospitalization.
Other viruses were identified in 20% of RSV+ infants, and at similar rates in both RSV types (P = .45). RSV/rhinovirus (16%) followed by RSV/adenovirus (2%) were the most frequent combinations. Sensitivity analyses excluding infants with viral codetection showed comparable results, and confirmed that genomic loads were higher in infants with RSV A versus RSV B, but disease severity was similar irrespective of the viral type (data not shown).
Patient Characteristics According to RSV Genotypes
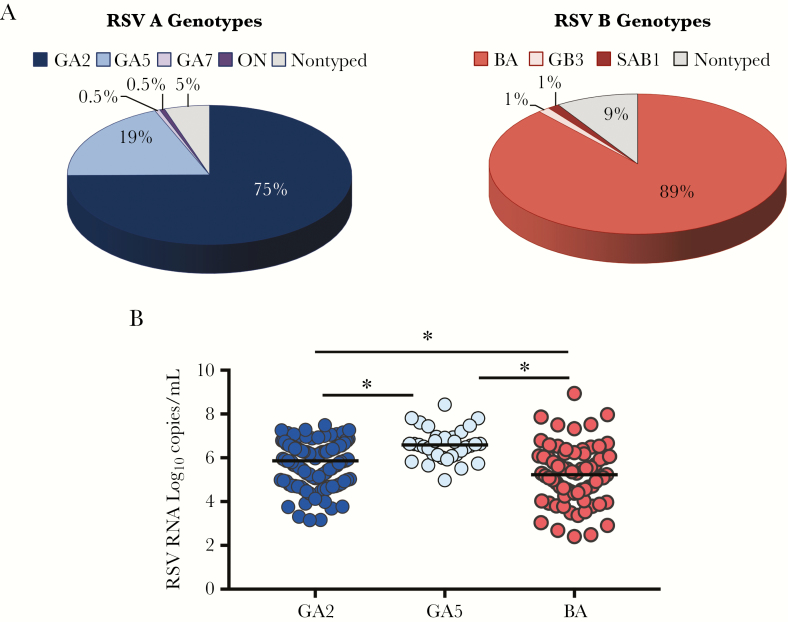
We next analyzed the variability of RSV genotypes over time (Figure 2B), and determined whether the different genotypes and viral loads from a specific genotype were associated with disease severity. Overall, there was a greater variability among RSV A isolates. During the initial years of the study (2004 to 2006) the predominant RSV A genotype was GA5, which was replaced by the GA2 genotype in 2007. RSV A GA7 was identified in one patient during the first study season, and the newly identified ON1 genotype also in 1 patient during the 2010–11 season. With respect to the 78 RSV B types, 69 (88%) corresponded to the BA genotype, which circulated at similar rates across the 5 study seasons. Genotypes B/SAB1 and B/GB3 were each identified in 1 patient during the 2004–2005 and 2005–2006 seasons, respectively. Nine (5%) RSV A isolates and 7 (9%) RSV B strains were not typed due to insufficient RNA (Figure 3A).
Figure 3.
Respiratory syncytial virus (RSV) distribution by genotypes and genotype loads. A, Pie graphs represent the genetic variability within RSV A and B isolates. Within RSV A genotypes, GA2 was the most commonly identified in 75% of children, followed by GA5 in 19% of patients. Of the 11 remaining RSV A genotypes, 1 corresponded to RSV GA7, 1 to the newly identified ON1 genotype, and 5% were not typed. Within the RSV B types, 88% corresponded to the BA genotype, 1 to RSV GB3, 1 to SAB1, and 9% were not typed. B, RSV genomic loads (vertical [y] axis) according to the 3 main genotypes BA (light red), GA2 (dark blue), and GA5 (light blue). Analyses performed using Kruskal–Wallis test following Benjamini–Hochberg to adjust for multiple comparisons. * Adjusted P < .05 was considered significantly different.
Demographic characteristics and disease severity parameters according to the 3 most frequent genotypes (GA2, GA5, and BA) are shown in Table 2. We found significant differences in terms of race/ethnicity with a higher proportion of infections due to the GA2 genotype in white children, while GA5 was most commonly identified in Hispanic children, possibly associated with the enrollment site (Ohio versus Texas). Infants infected with RSV BA had symptoms for a longer time before hospitalization and also had lower viral loads compared with those infected with the GA2 or GA5 genotypes (Figure 3B). Nevertheless, disease severity assessed by the CDSS, administration of supplemental O2, PICU admission, and duration of hospital stay were not influenced by the viral genotype or genotype-specific viral load.
Table 2.
Demographic, Clinical Data, and Viral Loads According to the Most Frequent Respiratory Syncytial Virus Genotypes
| GA2 (n = 131) |
GA5
(n = 33) |
BA
(n = 69) |
P value | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age, months | 2.1 [1.2–4.1] |
2.4 [0.8–5.7] |
2.2 [1.1–3.4] |
.85 |
| Gender, n (%) | ||||
| Male | 73 (56) | 20 (61) | 36 (52) | .71 |
| Female | 58 (42) | 13 (39) | 33 (48) | |
| Race/ethnicity, n (%) | ||||
| White | 97 (74) | 11 (33) | 40 (58) | |
| Black | 20 (15) | 6 (18) | 10 (15) | <.01 |
| Hispanic | 10 (8) | 15 (46) | 12 (17) | |
| Other | 4 (3) | 1 (3) | 7 (10) | |
| Breastfeeding (%) | 29 | 33 | 39 | .39 |
| Second hand smoking (%) | 42 | 21 | 29 | .06 |
| Daycare (%) | 20 | 14 | 9 | .44 |
| Siblings (%) | 79 | 93 | 86 | .08 |
| Disease severity | ||||
| Days of symptoms | 4 [3–6] | 4 [2–5] | 5 [3–7] | .04 |
| O2 requirement | ||||
| n (%) | 100 (76) | 19 (61)a | 47 (68) | .17 |
| Duration of oxygen (days) | 1.5 [0.1–4] | 1 [0–2]) | 1 [0–3] | .10 |
| PICU, n (%) | 31 (24) | 4 (12) | 15 (22) | .35 |
| CDSS | 5 [3–8] | 5 [3–7] | 5 [3–8] | .70 |
| LOS, days | 3 [2–5] | 3 [2–8.5] | 3 [2–6] | .49 |
| Viral load, log10 copies/mL | 5.86 [5.03–6.45] |
6.59 [6.10–6.94] |
5.23 [4.32–6.13] |
<.01 |
Continuous variables were analyzed using the Kruskall–Wallis tests and categorical variables using Fisher’s exact or Chi-square test. Data are reported as medians, 25%–75% interquartile range.
Abbreviations: PICU, pediatric intensive care unit; CDSS, clinical disease severity score; LOS, length of stay.
aFor GA5 genotype O2 administration was not documented in two patients.
Bold highlights the significant.
Host Transcriptional Profiles According to RSV Types and Genotypes
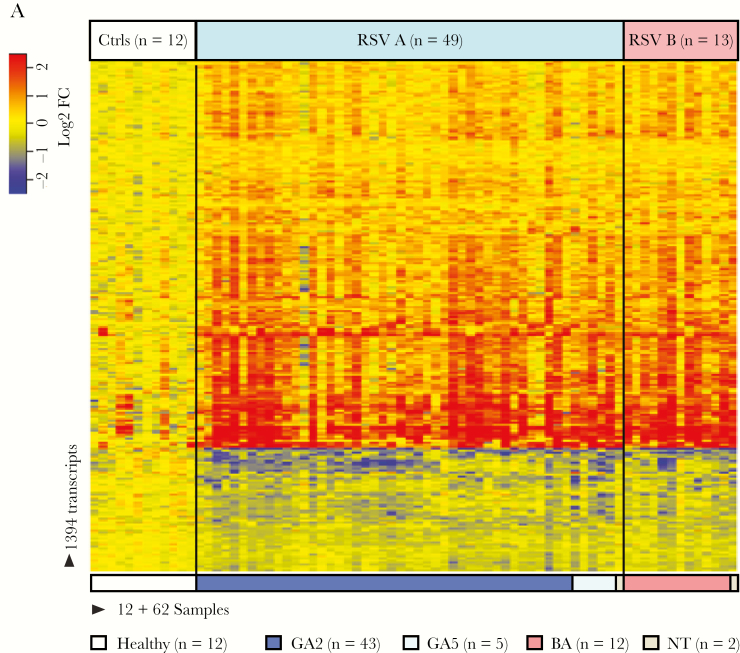
To define the host systemic immune response according to the different RSV types and genotypes, blood transcriptional profiles were analyzed in a subset of infants with RSV bronchiolitis with available samples. These 62 infants were enrolled during the last study season (2010–2011) along with 12 healthy asymptomatic age-matched controls (Supplementary Table S2). Of the 62 infants with RSV LRTI analyzed, 49 (79%) were infected with RSV A (GA2 n = 43, GA5 n = 5, nontyped n = 1) and 13 (21%) with RSV B (BA n = 12, nontyped n = 1).
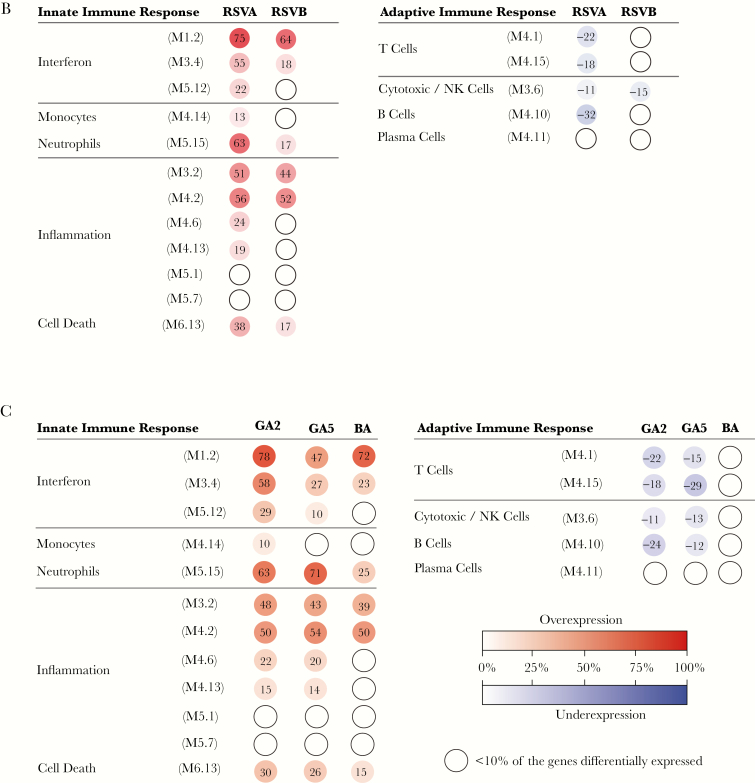
Statistical group comparisons (Benjamini–Hochberg corrected FDR <0.01 and ≥1.25 fold change) using linear mixed models identified 1234 transcripts differentially expressed between 49 infants with RSV A and healthy controls, and 640 transcripts between 13 infants with RSV B and controls. These transcripts were combined using a Venn diagram that yielded a total of 1394 transcripts identified in RSV A or B infection (Figure 4A). To further characterize the biological significance of the RSV A and B profiles and the function of the differentially expressed genes, we applied a modular analytical tool [33]. Modular profiles were derived separately for RSV A and B infection in comparison with healthy age-matched controls (Figure 4B). Infants with RSV A infection demonstrated significantly greater overexpression of interferon, inflammation, and neutrophil modules (adjusted (a) P values < .01) compared with RSV B, while no significant differences were observed in adaptive immunity-related modules. We followed a similar approach based on the 3 major genotypes identified: GA2, GA5, and BA (Figure 4C). Infants infected with the GA5 genotype demonstrated significantly lower overexpression of interferon-related modules, and also displayed greater activation of neutrophil related transcripts compared to the GA2 and BA genotypes (aP values < .001).
Figure 4.
Host transcriptional profiles according to respiratory syncytial virus (RSV) types and genotypes. A, class comparisons (Benjamini–Hochberg corrected FDR <0.01 and ≥1.25 fold change) using linear mixed models between infants with RSV A (n = 49) versus age-matched controls (n = 12), and infants with RSV B (n = 13) versus the same controls, identified 1394 transcripts that were present in either of these infections. Transcripts are organized in a heatmap format where each row represents a single transcript and each column represents a patient sample. Red indicates overexpression and blue underexpression of a transcript compared to the median expression of healthy controls (yellow). Color bars above and below the heatmap indicate the patient groups, controls, or RSV types (A or B), and genotypes (GA2, GA5, and BA). One RSV A sample and one RSV B sample were not typed due to insufficient RNA (white). B and C, To characterize biological functions of the differentially expressed genes, we used a modular-based analysis. Analysis was performed separately for (B) infants with RSV A and RSV B compared to healthy controls and (C) the 3 main genotypes: GA2 (n = 43), GA5 (n = 5), and BA (n = 12). The intensity of the modules (dots) indicates the proportion of overexpressed (red) or underexpressed (blue) transcripts within each module in relation to the healthy control baseline. Numeric values indicate the percentage of transcripts expressed in each specific module. Abbreviation: NK cell, natural killer cell.
Analyses of Independent Effects in Clinical Outcomes
Independent predictors of disease severity were analyzed using multivariable logistic regression analyses. We selected 2 clinical outcomes, length of hospital stay and PICU admission, as indicators of severe disease. Age, gender, the CDSS, which was dichotomized into mild (0–5) versus moderate–severe (6–15), RSV genomic loads, and the most relevant RSV genotypes (GA2, GA5, and BA) were included in the models as covariates.
Younger age, a CDSS severity score >5 and the RSVA/GA5 genotype were significantly associated with prolonged length of hospital stay. On the other hand, only the CDSS >5 was a predictor of admission to the PICU (Table 3). Specific genotype viral loads were not significantly associated with the clinical outcomes evaluated (data not shown).
Table 3.
Host and Viral Factors as Independent Predictors of Respiratory Syncytial Virus Disease Severity
| Length of Stay RR [95% CI] |
P value | PICU Admission OR [95% CI] |
P value | |
|---|---|---|---|---|
| Age | 1.04 [1.01–1.06] | .004 | 1.07 [0.95–1.20] | .25 |
| Gender (male) | 0.84 [0.70–1.01] | .06 | 1.82 [0.83–3.97] | .13 |
| Severe disease (CDSS 6–15) | 1.92 [1.61–2.29] | <.0001 | 30.99 [10.43–92.05] | <.0001 |
| Genotype BA | 1.20 [0.98–1.47] | .07 | 0.83 [0.34–2.02] | .61 |
| Genotype GA5 | 1.44 [1.11–1.87] | .006 | 0.41 [0.11–1.55] | .24 |
| Viral load, log10 copies/mL | 1.00 [0.92–1.09] | .85 | 0.58 [0.58–1.23] | .38 |
Association of predictors with length of stay is displayed using relative risk (RR) and 95% confidence intervals (CI), and PICU admission using odds ratios (OR) and 95% CI. Reference value for genotypes is GA2. The CDSS ranged from 0 to 15, with higher scores indicating more severe disease. The CDSS was dichotomized into severe (6–15) versus mild (0–5). OR for this covariate is displayed in relation to mild disease.
Abbreviations: PICU, pediatric intensive care unit; CDSS, clinical disease severity score.
Bold highlights the significant.
DISCUSSION
The main purpose of this study was to define the variability of RSV A and B genotypes over time, to assess whether RSV types and genotypes and viral loads were associated with disease severity, and to determine whether the RSV types and genotypes identified induced distinct host immune profiles. We found that both RSV types cocirculated during each of the seasons analyzed, but RSV A consistently predominated over RSV B infections. The main RSV B genotype, BA, was identified at similar rates across the 5 study years. However, RSV A/GA5, which was the predominant RSV A genotype initially, was replaced by GA2 during the last 3 study seasons. Although we did not observe an overall association between RSV genomic loads and disease severity, infection with RSV A/GA5 was independently associated with worse clinical outcomes, as well as a distinctive host immune profile characterized by greater overexpression of neutrophil genes and less activation of interferon-related genes.
Previous studies have evaluated the association between RSV loads and disease severity with conflicting results [10, 19, 34–40]. While some studies found significant correlations between viral loads and clinical severity, others failed to identify such associations. A multicenter prospective cohort study that included more than 1500 children <2 years of age hospitalized with RSV bronchiolitis found that children with higher genomic loads had a higher risk of PICU admission, assisted ventilation (CPAP or intubation), and longer hospital stay, despite having similar clinical disease severity at the time of presentation [38]. On the other hand, 2 large studies recently conducted in children <5 years of age with LRTI/pneumonia in developing countries, did not show significant associations between RSV genomic loads and any of the clinical outcomes evaluated, including PICU admission, mechanical ventilation, length of stay, or death [39, 41]. In the present study, although genomic loads were higher in infants with RSV A versus RSV B infection, we did not find significant associations between overall RSV genomic loads or specific genotype loads and more severe clinical illness. This may be attributed to the smaller sample size of our study, and possibly to the fact that viral replication is a dynamic process and although patients were enrolled and sampled within 24 hours of hospitalization, they were hospitalized at different times during the course of their illness.
The association between RSV types (A or B) and disease severity has also been studied. Although some studies showed worse disease severity among children with RSV A infections, other studies have not supported those findings [2–8, 42]. In the present study, the clinical presentation and overall disease severity of children hospitalized with RSV A or B infections were similar. The different results with regards to viral loads and RSV types between previous studies and the present one could be attributed to differences in study design, such as the indicators of disease severity analyzed, the patient populations included (because some studies included infants with underlying diseases or were focused on the outpatient setting), the assays used to measure RSV loads (plaque assay, reflective of infectious load, versus semiquantitative or quantitative real time PCR, which measures genomic loads), or the differences in duration of symptoms at the time of study enrollment and sampling.
Studies to determine the genetic diversity of RSV strains over time have been conducted worldwide. In the majority of those studies, RSV detection and genotyping was performed either as part of a respiratory viral surveillance program, usually targeting children <5 years of age or older, including small sample sizes, or by analyzing respiratory samples already collected per standard of care, which may underestimate the burden of RSV genotypes in infants and young children with RSV infection [9, 11, 12, 16, 42–48]. In the present study we followed a different approach, by prospectively enrolling a large population of previously healthy infants hospitalized with RSV bronchiolitis and analyzing in detail the association between RSV types, RSV genotypes, and genotype-specific viral loads and disease severity over several respiratory seasons. Nevertheless, in agreement with those studies, we observed a replacement of the predominant GA5 genotype by GA2 from 2004 through 2010, a consistent proportion of BA genotypes, and the identification of ON1 genotype during the last study season 2010–2011. A summary of the different studies detailing the circulation and replacement of genotypes over time in different countries is summarized in Supplementary Table S1.
Limited data derived from in vitro and animal models suggest that, depending on the infecting RSV strain, host responses may be different. A549 cells infected with 2 different RSV B clinical strains showed differences in interleukin-6 (IL-6) production [17], and on the other hand infection of well-differentiated pediatric bronchial epithelial cells showed that the GA5 strain was associated with greater epithelial sloughing and goblet cell hyperplasia than the laboratory-adapted GA1 strain [49]. In mice, greater IL-13 production, airway mucin expression, and tachypnea were observed after inoculation with different RSV A clinical isolates [50]. We took a different approach to determine whether RSV types or genotypes were associated with distinct host responses, and analyzed in a subset of infants the global host systemic immune response by RNA transcriptional profiling. Compared with the GA2 or BA genotypes, infants infected with RSV GA5 showed decreased induction of interferon expression, while there was enhanced overexpression of neutrophil-related genes. Moreover, these infants also displayed greater clinical severity as shown by their longer length of hospital stay, adjusted for all other covariates. Taken together, these data suggest the possibility of RSV strain-specific differences in disease pathogenesis that contribute to clinical severity.
Our study has limitations. As mentioned above, nasopharyngeal aspirates were collected at a single time point, and thus we could not assess viral load dynamics in relation to disease severity. However, 2 of our main objectives were to determine whether RSV genotypes and genotype-specific genomic loads measured at enrollment were associated with enhanced disease severity. Our study was conducted in 2 centers in the United States, included a convenience sample of hospitalized children, and thus may not be generalizable. Nevertheless, we enrolled a relatively large cohort of infants and young children (median age of 2.1 months) without underlying diseases, of different backgrounds, and over 5 respiratory seasons. While the CDSS was consistently associated with the 2 major clinical outcomes evaluated, infection with the RSV A/GA5 genotype was significantly associated with prolonged length of stay, but not with PICU admission. This particular genotype and other viral proteins may account for differences in disease severity, by modulating the host immune response, which deserves further studies.
In conclusion, RSV types A and B cocirculated during each respiratory season without a clear association between RSV types, genomic loads, or specific genotype loads and disease severity. On the other hand, the RSV A genotype GA5, which predominated during the initial study years, was associated with distinct host immune profiles and independently predicted prolonged length of hospitalization, an important indicator of disease severity. Further studies, including sequential sampling, sequencing other viral genes, and enrolling infants representative of the whole disease spectrum, are warranted.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Presented in parts: This work was presented in part at the Infectious Diseases Society of America 48th Meeting, October 2010, Vancouver, BC, Canada.
Notes
Acknowledgments. We would like to thank all the team at Clinical Research for their extraordinary efforts enrolling our patients, Shama Khokhar and Sara Mertz for help running the viral PCRs, Nicole Baldwin for her help curating the transcriptomic data, and especially to our patients and their families for agreeing to participate in this study.
Financial support. This work was supported by NIH grants AI112524 and intramural grants to A. M., O. R., and M. E. P.
Potential conflicts of interest. A. M., O. R., and M. E. P. have received research grants from Janssen. A. M. has received fees for participation in advisory boards from Janssen and lectures from Abbvie and Novartis. O. R. has received fees for participation in advisory boards from Abbvie, HuMabs, Janssen, Medimmune, and Regeneron, and lectures from Abbvie. M. E. P. has received research grants from Agilvax, and the CF Foundation, fees for lectures from Pfizer, and participation in an advisory board from ReViral. P. A. P. has received research grants from Novavax, Gilead, and Janssen and has served on Scientific Advisory Board for Novavax, MedImmune, Ablynx, and LFB Biotechnologies. Those grants and fees were not related to the research described in this manuscript. All other authors reported no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Leader S, Kohlhase K. Respiratory syncytial virus-coded pediatric hospitalizations, 1997 to 1999. Pediatr Infect Dis J 2002; 21:629–32. [DOI] [PubMed] [Google Scholar]
- 2. McConnochie KM, Hall CB, Walsh EE, Roghmann KJ. Variation in severity of respiratory syncytial virus infections with subtype. J Pediatr 1990; 117:52–62. [DOI] [PubMed] [Google Scholar]
- 3. Imaz MS, Sequeira MD, Videla C et al. . Clinical and epidemiologic characteristics of respiratory syncytial virus subgroups A and B infections in Santa Fe, Argentina. J Med Virol 2000; 61:76–80. [DOI] [PubMed] [Google Scholar]
- 4. Walsh EE, McConnochie KM, Long CE, Hall CB. Severity of respiratory syncytial virus infection is related to virus strain. J Infect Dis 1997; 175:814–20. [DOI] [PubMed] [Google Scholar]
- 5. Jafri HS, Wu X, Makari D, Henrickson KJ. Distribution of respiratory syncytial virus subtypes A and B among infants presenting to the emergency department with lower respiratory tract infection or apnea. Pediatr Infect Dis J 2013; 32:335–40. [DOI] [PubMed] [Google Scholar]
- 6. Brouard J, Freymuth F, Constantini S, Petitjean J, de Schrevel G, Duhamel JF. Prevalence and clinical aspects of A and B subgroups of respiratory syncytial virus infection. Observation of 8 consecutive epidemics between 1982 and 1990. Arch Fr Pediatr 1993; 50:639–43. [PubMed] [Google Scholar]
- 7.McIntosh ED, De Silva LM, Oates RK. Clinical severity of respiratory syncytial virus group A and B infection in Sydney, Australia. Pediatr Infect Dis J 1993; 12:815–9. [DOI] [PubMed] [Google Scholar]
- 8. Kneyber MC, Brandenburg AH, Rothbarth PH, de Groot R, Ott A, van Steensel-Moll HA. Relationship between clinical severity of respiratory syncytial virus infection and subtype. Arch Dis Child 1996; 75:137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esposito S, Piralla A, Zampiero A et al. . Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in Northern Italy in five consecutive winter seasons. PLoS One 2015; 10:e0129369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fodha I, Vabret A, Ghedira L et al. . Respiratory syncytial virus infections in hospitalized infants: association between viral load, virus subgroup, and disease severity. J Med Virol 2007; 79:1951–8. [DOI] [PubMed] [Google Scholar]
- 11. Gilca R, De Serres G, Tremblay M et al. . Distribution and clinical impact of human respiratory syncytial virus genotypes in hospitalized children over 2 winter seasons. J Infect Dis 2006; 193:54–8. [DOI] [PubMed] [Google Scholar]
- 12. Martinello RA, Chen MD, Weibel C, Kahn JS. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis 2002; 186:839–42. [DOI] [PubMed] [Google Scholar]
- 13. Papadopoulos NG, Gourgiotis D, Javadyan A et al. . Does respiratory syncytial virus subtype influences the severity of acute bronchiolitis in hospitalized infants?Respir Med 2004; 98:879–82. [DOI] [PubMed] [Google Scholar]
- 14. Smyth RL, Mobbs KJ, O’Hea U, Ashby D, Hart CA. Respiratory syncytial virus bronchiolitis: disease severity, interleukin-8, and virus genotype. Pediatr Pulmonol 2002; 33:339–46. [DOI] [PubMed] [Google Scholar]
- 15. .Otieno JR, Kamau EM, Agoti CN et al. . Spread and evolution of respiratory syncytial virus A genotype ON1, Coastal Kenya, 2010–2015. Emerg Infect Dis 2017; 23:264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Espinosa Y, San Martin C, Torres AA et al. . Genomic loads and genotypes of respiratory syncytial virus: viral factors during lower respiratory tract infection in Chilean hospitalized infants. Int J Mol Sci 2017; 18:E654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levitz R, Wattier R, Phillips P et al. . Induction of IL-6 and CCL5 (RANTES) in human respiratory epithelial (A549) cells by clinical isolates of respiratory syncytial virus is strain specific. Virol J 2012; 9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. American Academy of Pediatrics Subcommittee on Diagnosis and Management of Bronchiolitis. Diagnosis and management of bronchiolitis. Pediatrics 2006; 118:1774–93. [DOI] [PubMed] [Google Scholar]
- 19. Sheeran P, Jafri H, Carubelli C et al. . Elevated cytokine concentrations in the nasopharyngeal and tracheal secretions of children with respiratory syncytial virus disease. Pediatr Infect Dis J 1999; 18:115–22. [DOI] [PubMed] [Google Scholar]
- 20. Gill MA, Long K, Kwon T et al. . Differential recruitment of dendritic cells and monocytes to respiratory mucosal sites in children with influenza virus or respiratory syncytial virus infection. J Infect Dis 2008; 198:1667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mella C, Suarez-Arrabal MC, Lopez S et al. . Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis 2013; 207:564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. García C, Soriano-Fallas A, Lozano J et al. . Decreased innate immune cytokine responses correlate with disease severity in children with respiratory syncytial virus and human rhinovirus bronchiolitis. Pediatr Infect Dis J 2012; 31:86–9. [DOI] [PubMed] [Google Scholar]
- 23. Suárez-Arrabal MC, Mella C, Lopez SM et al. . Nasopharyngeal bacterial burden and antibiotics: Influence on inflammatory markers and disease severity in infants with respiratory syncytial virus bronchiolitis. J Infect 2015; 71:458–69. [DOI] [PubMed] [Google Scholar]
- 24. Nolte FS, Marshall DJ, Rasberry C et al. . MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J Clin Microbiol 2007; 45:2779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mejias A, Dimo B, Suarez NM et al. . Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinonen S, Jartti T, Garcia C et al. . Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med 2016; 193:772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peret TC, Hall CB, Hammond GW et al. . Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis 2000; 181:1891–6. [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–9. [DOI] [PubMed] [Google Scholar]
- 29. Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 2004; 5:150–63. [DOI] [PubMed] [Google Scholar]
- 30. Tapia LI, Shaw CA, Aideyan LO et al. . Gene sequence variability of the three surface proteins of human respiratory syncytial virus (HRSV) in Texas. PLoS One 2014; 9:e90786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berry MP, Graham CM, McNab FW et al. . An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Banchereau R, Jordan-Villegas A, Ardura M et al. . Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One 2012; 7:e34390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaussabel D, Quinn C, Shen J et al. . A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 2008; 29:150–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeVincenzo JP, El Saleeby CM, Bush AJ. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis 2005; 191:1861–8. [DOI] [PubMed] [Google Scholar]
- 35. Houben ML, Coenjaerts FE, Rossen JW et al. . Disease severity and viral load are correlated in infants with primary respiratory syncytial virus infection in the community. J Med Virol 2010; 82:1266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall CB, Douglas RG Jr, Geiman JM. Quantitative shedding patterns of respiratory syncytial virus in infants. J Infect Dis 1975; 132:151–6. [DOI] [PubMed] [Google Scholar]
- 37. Wright PF, Gruber WC, Peters M et al. . Illness severity, viral shedding, and antibody responses in infants hospitalized with bronchiolitis caused by respiratory syncytial virus. J Infect Dis 2002; 185:1011–8. [DOI] [PubMed] [Google Scholar]
- 38. Hasegawa K, Jartti T, Mansbach JM et al. . Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis 2015; 211:1550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazur NI, Bont L, Cohen AL et al. . Severity of respiratory syncytial virus lower respiratory tract infection with viral coinfection in HIV-uninfected children. Clin Infect Dis 2016; 64:443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scagnolari C, Midulla F, Selvaggi C et al. . Evaluation of viral load in infants hospitalized with bronchiolitis caused by respiratory syncytial virus. Med Microbiol Immunol 2012; 201:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feikin DR, Fu W, Park DE et al. ; PERCH Study Group Is higher viral load in the upper respiratory tract associated with severe pneumonia? Findings from the PERCH study. Clin Infect Dis 2017; 64:337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoshihara K, Le MN, Okamoto M et al. . Association of RSV-A ON1 genotype with increased pediatric acute lower respiratory tract infection in Vietnam. Sci Rep 2016; 6:27856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luchsinger V, Ampuero S, Palomino MA et al. . Comparison of virological profiles of respiratory syncytial virus and rhinovirus in acute lower tract respiratory infections in very young Chilean infants, according to their clinical outcome. J Clin Virol 2014; 61:138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panayiotou C, Richter J, Koliou M, Kalogirou N, Georgiou E, Christodoulou C. Epidemiology of respiratory syncytial virus in children in Cyprus during three consecutive winter seasons (2010–2013): age distribution, seasonality and association between prevalent genotypes and disease severity. Epidemiol Infect 2014; 142:2406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tran DN, Pham TM, Ha MT et al. . Molecular epidemiology and disease severity of human respiratory syncytial virus in Vietnam. PLoS One 2013; 8:e45436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venter M, Collinson M, Schoub BD. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: comparison of viruses and genotypes responsible for different disease manifestations. J Med Virol 2002; 68:452–61. [DOI] [PubMed] [Google Scholar]
- 47. Otieno JR, Agoti CN, Gitahi CW et al. . Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in Coastal Kenya. J Virol 2016; 90:4990–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hause AM, Henke DM, Avadhanula V, Shaw CA, Tapia LI, Piedra PA. Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B. PLoS One 2017; 12:e0175792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Villenave R, Thavagnanam S, Sarlang S et al. . In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc Natl Acad Sci U S A 2012; 109:5040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stokes KL, Chi MH, Sakamoto K et al. . Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 2011; 85:5782–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.