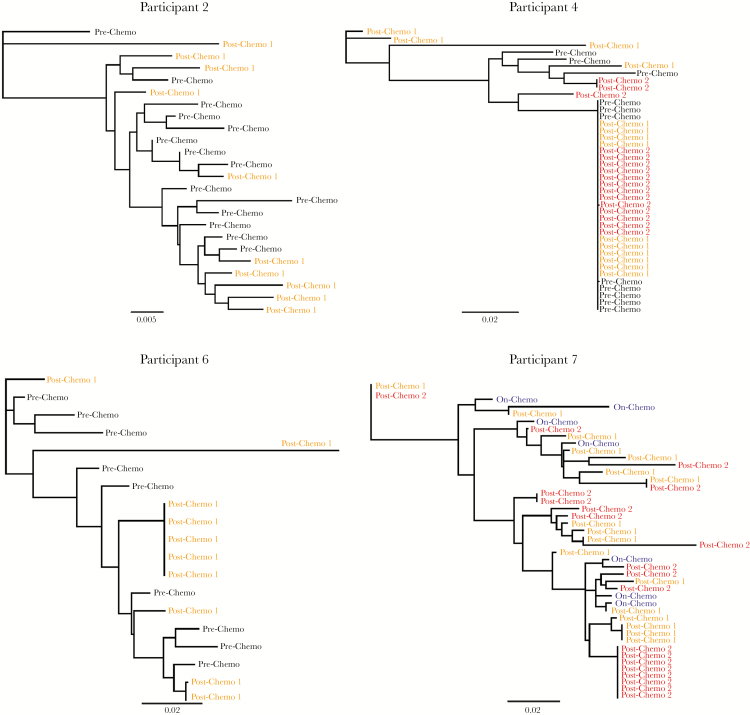
Figure 3.
Maximum likelihood phylogenetic trees of single-genome human immunodeficiency virus type 1 (HIV-1) envelope DNA sequences from pre- or on-chemotherapy and postchemotherapy CD4+ T cells in patients with lymphoma on stable antiretroviral therapy throughout chemotherapy. Oligoclonal clustering of sequences was observed only following completion of chemotherapy in participants 4 and 6, with an increase in the number of near-identical envelope sequences following chemotherapy in participant 7. Phylogenetic trees were generated using bootstrapped, generalized time-reversible models. Bars represent genetic distance. Mean genetic distances for each participant time point are as follows: participant 2 prechemotherapy D = 0.019, postchemotherapy D = 0.032; participant 4 prechemotherapy D = 0.015, first postchemotherapy D = 0.032, second postchemotherapy D = 0.012; participant 6 prechemotherapy D = 0.018, postchemotherapy D = 0.019; participant 7 first on-chemotherapy D = 0.046, postchemotherapy D = 0.033. When excluding the single postchemotherapy outlier sequence from participant 6, the prechemotherapy D = 0.024 and postchemotherapy D = 0.013.