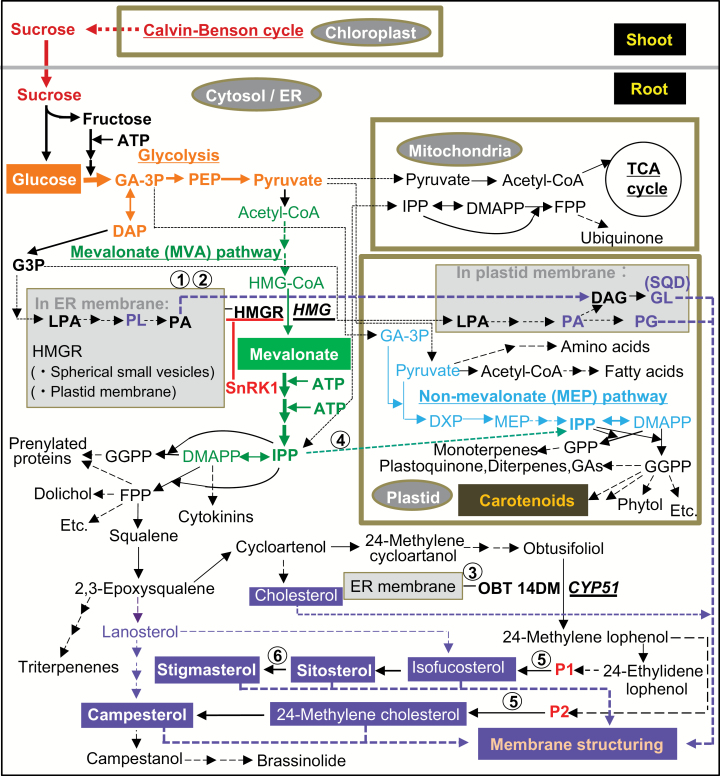
Fig. 7.
Model for biosynthesis of sterols and other isoprenoids in rice roots. Two independent pathways produce IPP, the precursor of all isoprenoids: the cytosolic acetate/mevalonate (MVA) pathway and the plastidial non-MVA methylerythritol 4-phosphate (MEP) pathway. Figure was compiled by reference to Fig. 1 of Fujioka et al. (1997), Fig. 1 of Rodríguez-Concepción et al. (2004), and Fig. 1 of Wang et al. (2012). Thicker arrows or bold letters in cytosol indicate presumed high concentrations of metabolites under dark conditions with mevalonate and glucose supplied (this study; see Discussion). Glycolysis, MVA, and MEP pathways are shown in orange, green, and blue letters, respectively. Purple dashed arrow shows biosynthetic pathway to phytosterol via lanosterol in dicots. ①, localization of HMGR at endoplasmic reticulum (ER) membrane and partially at spherical small vesicles (Leivar et al. 2005); ②, localization of HMGR at plastid membrane (Wong et al. 1982); ③, localization site of OBT 14DM at ER membrane (Bak et al. 1997); ④, greater inhibition of cytosolic IPP partitioning from the cytosol to the plastid by Al treatment in Al-sensitive Ko under DMG than under L; ⑤, Δ5,7-sterol Δ7-reductase encoded by DWARF5; ⑥, sterol-Δ22-desaturase encoded by CYP710A1; DAP, dihydroxyacetone phosphate; DMAPP, dimethylallyl diphosphate; DXP, 1-deoxy-D-xylulose-5-phosphate; FPP, farnesyl diphosphate; GA-3P, glyceraldehyde-3-phosphate; GAs, gibberellins; GGPP, geranylgeranyl diphosphate; G3P, glycerol 3-phosphate; GL, galactolipids; GPP, geranyl diphosphate; HMG, 3-hydroxy-3-methylglutaryl-CoA reductase encoding gene; HMGR, HMG reductase; IPP, isopentenyl diphosphate; LPA, lysophosphatidate; MEP, methylerythritol-4-phosphate; P1, stigmasta-5,7,Z-24(241)-trien-3β-ol; P2, ergosta-5,7,24(241)-trien-3β-ol; PEP, phosphoenol pyruvate; PG, phosphatidylglycerol; PL, phospholipids; SnRK1, sucrose non-fermenting 1-related kinase 1; SQD, sulfoquinovosyldiacylglycerol.