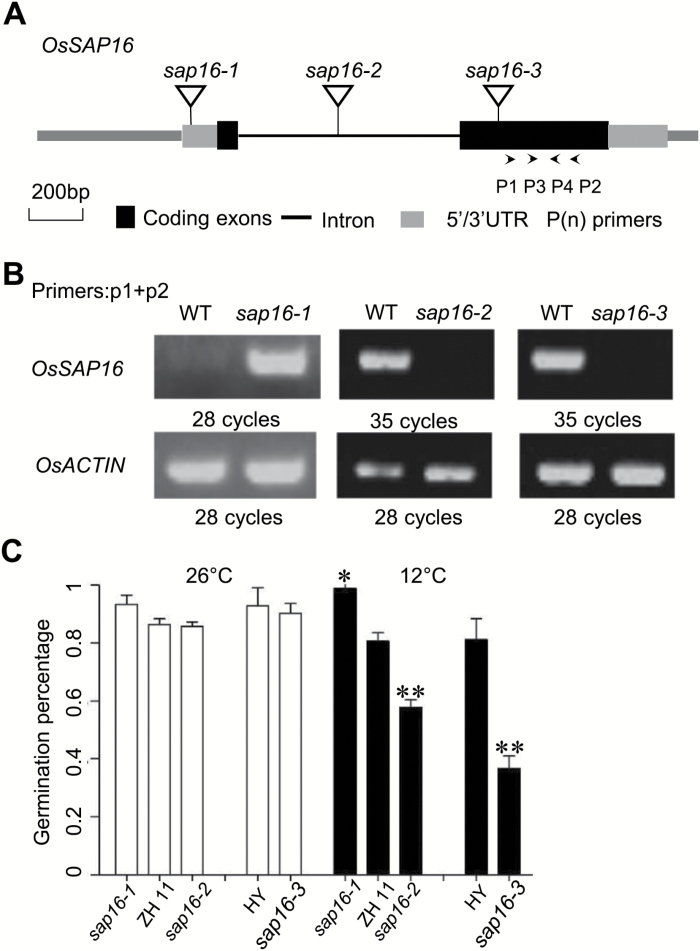
Fig. 4.
Identification of OsSAP16 as the causal gene for a LTG QTL. (A) Schematic presentation of the gene structure of OsSAP16 and T-DNA insertion sites in three OsSAP16 mutants (indicated by arrowheads). P1 and P2, and P3 and P4, are two pairs of primers used to amplify OsSAP16 cDNA for semiquantitative PCR analysis. (B) Semiquantitative RT-PCR analysis of OsSAP16 expression in mutants. The OsACTIN gene was used as a normalization control. Shown are the products from 28 cycles of amplification for OsACTIN in all samples, 28 cycles of OsSAP16 in sap16-1, and 35 cycles of OsSAP16 for sap16-2 and sap16-3. (C) Germination of wild-type Zhonghua11 (ZH11), Hwayoung (HY), and the three T-DNA insertion mutants of OsSAP16 under 12 °C and 26 °C growth conditions. sap16-1 in ZH11 is an overexpression mutant, and sap16-2 in ZH11 and sap16-3 in HY are loss-of-function mutants. Seeds were incubated for 3 days at 26 °C and for 11 days at 12 °C. Data are the means±SD from three replicates. Differences between mutants and wild types were analyzed by Welch’s t-test. *P<0.05, **P<0.01.