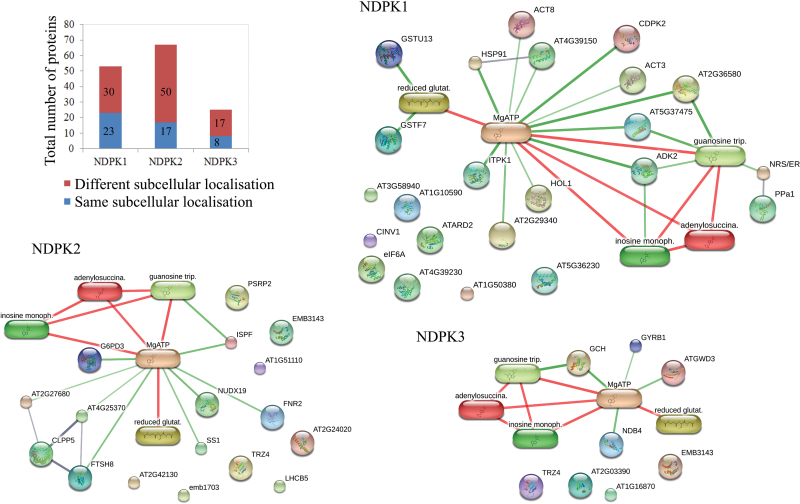
Fig. 3.
Proteins and small-molecule ligands pulled-down with NDPKs. The total number of protein partners co-purified with NDPK1–NDPK3 based on absence from the negative list but presence in the C- and N-tagged samples (see also Supplementary Tables S9 and S10). Blue indicates proteins that passed the subcellular localization filter: 23, 17, and 8 for NDPK1, 2, and 3, respectively. These, together with metabolites co-purified with NDPKs (glutathione, IMP, and adenylosuccinate) and nucleotide triphosphates directly related to NDPK function (ATP and GTP) were used to query the Stich database for protein–protein and protein–metabolite complexes. We used a minimum required interaction score of 0.5 and restricted our search to (i) experimentally, (ii) database-, and (ii) literature-reported associations. Subcellular protein localization was retrieved from the SUBA database (Tanz et al., 2012).