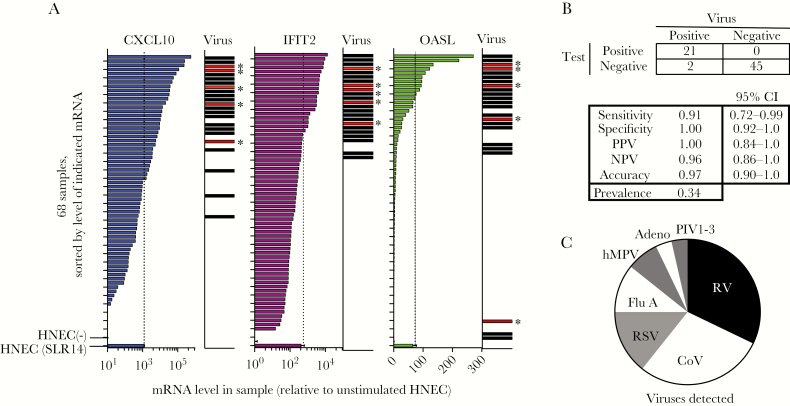
Figure 2.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) test for 3 host mRNAs and correlation with respiratory virus detection in 68 nasopharyngeal swabs (Study 1). A, Patient cells were pelleted from the transport medium containing patient nasopharyngeal swabs, RNA was isolated, and RT-qPCR was performed for β-actin and 3 mRNAs associated with the antiviral response as identified in Figure 1 (OASL, IFIT2, CXCL10). Plots show mRNA levels for 68 samples relative to the mRNA level in resting human nasal epithelial cells (HNEC (−)), sorted by mRNA level for each transcript. Biomarker mRNA was normalized to β-actin mRNA in the same sample. Each mRNA biomarker was scored positive if level was above the level observed in SLR14-stimulated HNEC, indicated by the dotted line and lowest bar on each graph (HNEC bars show mean and SD of 3 replicates). mRNA levels were normalized to the level of β-actin mRNA in each sample. Bars in virus column indicate samples that tested positive for respiratory virus by qPCR. Bars marked with asterisk indicate the 5 samples that were negative on the initial virology test panel but positive upon subsequent testing for CoV. B, Test performance of mRNA biomarker signature. Samples were scored as biomarker test positive if 2/3 mRNA biomarkers were above the cutoff (dotted lines). C, Pie chart shows relative abundance of the viruses detected in this sample set, representing 28 detections (23 virus positive samples with 5 codetections). Abbreviations: virus names are listed in Table 1; CoV, coronavirus OC43 (only CoV detected); NPV, negative predictive value; PPV, positive predictive value.