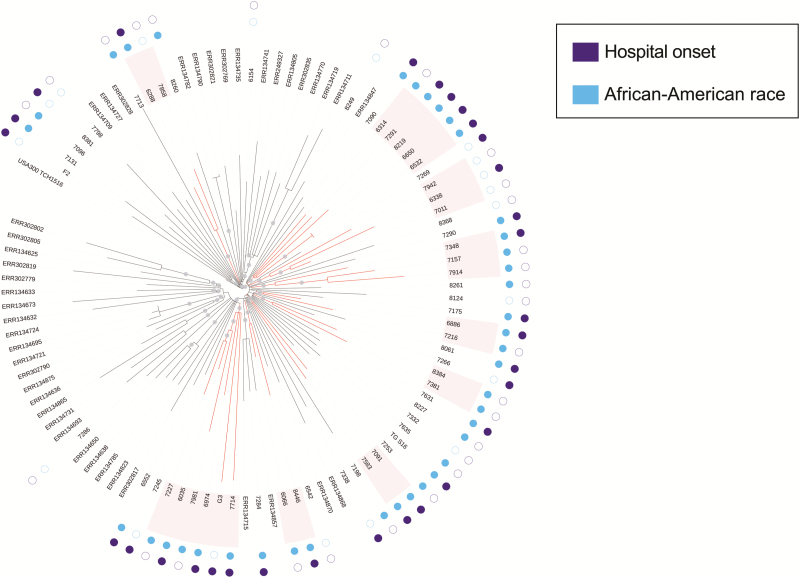
Figure 2.
Maximum-likelihood phylogeny of USA300 methicillin-resistant Staphylococcus aureus (MRSA) isolates from Chicago bloodstream infections and New York household surveillance. A phylogeny was constructed using genome-wide variants for the isolates from Figure 1, and an additional set of USA300 MRSA isolates collected during a previous study of New York households. Gray circles indicate splits with >90% bootstrap support. Red branches indicate subclades with >90% bootstrap support that contain only isolates from the current study, and are therefore deemed regional clusters. The inner ring of circles adjacent to the tip labels indicates whether the isolate was taken from an African-American individual (blue) and the outer circles indicate whether an isolates was designated hospital onset (purple).