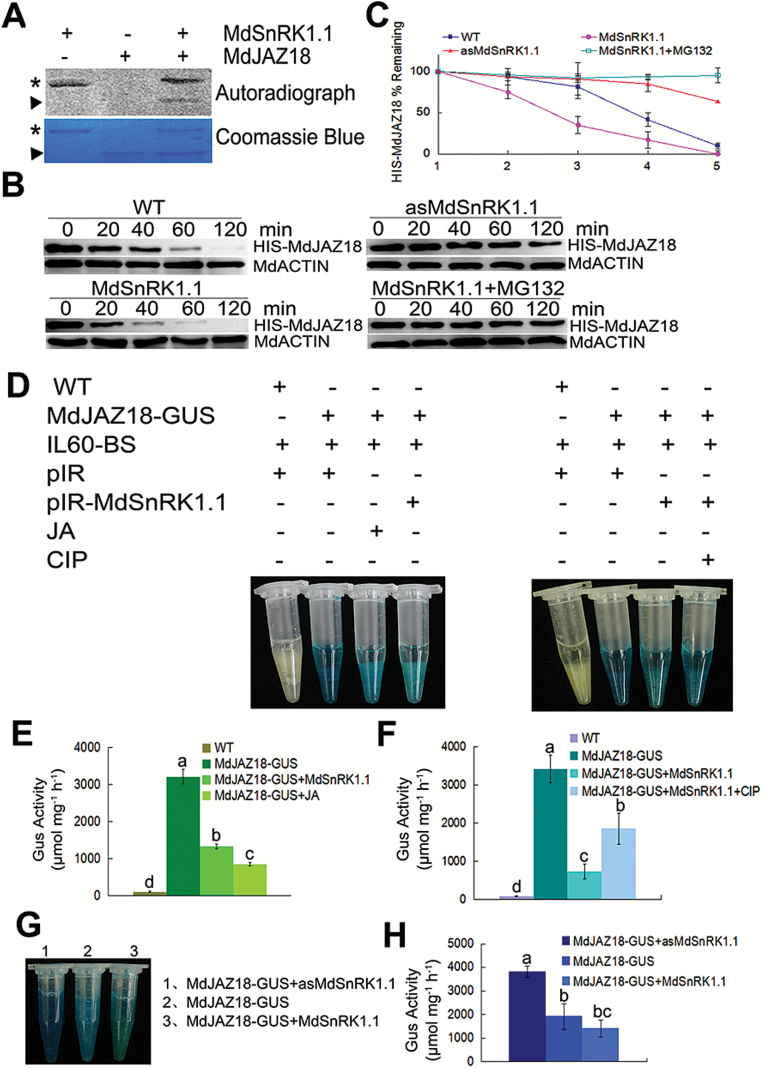
Fig. 4.
MdSnRK1.1 phosphorylates and degrades MdJAZ18 protein. (A) MdSnRK1.1 phosphorylates MdJAZ18 in vitro. The asterisk indicates the autophosphorylation of the purified HIS-MdSnRK1.1, while the triangle indicates the phosphorylation of the purified HIS-MdJAZ18 by HIS-MdSnRK1.1 in the autoradiogram. The asterisk and the triangle refer to protein loading of HIS-MdJAZ18 and HIS-MdSnRK1.1, respectively, in Coomassie blue staining. (B) MdJAZ18 stability is reduced by MdSnRK1.1 and the effect is inhibited by MG132. Total proteins extracted from WT, MdSnRK1.1, and asMdSnRK1.1 apple calli were incubated with the purified HIS-MdJAZ18 protein treated with or without MG132. The samples were harvested at the indicated time. MdACTIN was used as internal reference. (C) The half-life plot for in vitro HIS-MdJAZ18 degradation was analyzed from protein bands with Bio-Rad QuantityOne software. (D) MdSnRK1.1 degrades MdJAZ18, and CIP inhibits MdSnRK1.1-mediated degradation. GUS staining images of transiently expressed pIR-MdSnRK1.1 or pIR in MdJAZ18-GUS calli treated with or without JA or CIP. WT calli were used as a negative control. (E, F) Quantitative analysis of GUS activity in (D). (G) MdSnRK1.1 promoted the degradation of MdJAZ18 in vivo. GUS staining images of MdJAZ18–GUS+MdSnRK1.1, MdJAZ18–GUS+asMdSnRK1.1, and MdJAZ18–GUS calli. (H) Quantitative analysis of GUS activity in (G). In (C), (E), (F), and (H), error bars represent the SD. Statistical significance was calculated by the LSD test with DPS software, P<0.05.