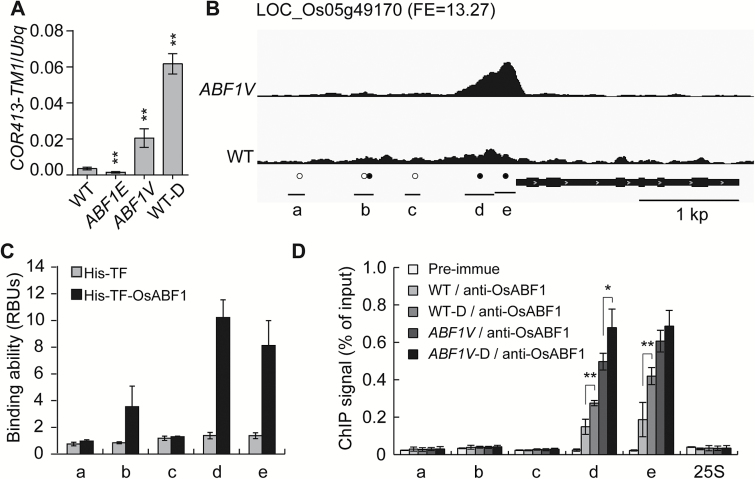
Fig. 5.
Identification of COR413-TM1 as a direct target of OsABF1. (A) Transcriptional analysis of COR413-TM1 in the WT, the ABF1E and ABF1V transgenic lines, and the WT-D treatment by quantitative reverse transcription PCR (qRT-PCR) with normalization to Ubiquitin-1 (Ubq). Mean values ±s.e.m. (standard error of the mean) are shown (Student’s t-tests, **P<0.01, n=3). (B) ABF1V binding profile in the promoter of COR413-TM1 in the ABF1V line and the WT ChIP-seq data. The bars at the bottom represent the distribution of DNA fragments containing the ACGTGG(T)C(A) motif or ACGT core, as indicated by black or white dots. (C) In vitro binding assay of His-TF-OsABF1 recombinant protein to the indicated DNA fragments, as shown in (B). His-TF recombinant protein was used as a negative control. (D) Verification of the OsABF1 direct binding sites in the COR413-TM1 promoter by ChIP-qPCR analysis. ChIP samples were prepared using the ABF1V line and the WT plants under normal conditions or under drought stress, and precipitated with anti-OsABF1 antibodies. The pre-immune serum was used as a negative control. Results of ChIP-qPCR were quantified by normalization of the immunoprecipitation signal with the corresponding input signal. The binding to 25S rDNA was used as a negative control. The means ±SD (n=3) are shown.